Program and confirmed speakers
Below you will find the list of confirmed speakers, the symposium programme, lectures abstracts and poster abstracts (updated all the time).
Offline (pdf) version of: program, lecture abstracts book and poster abstracts book.
Confirmed speakers:
- CEUM
- Lukáš Žídek
- CEITEC, Brno, Czech Republik
- Robert Konrat
- Max Perutz Labs, Vienna, Austria
- Piotr Garbacz
- Faculty of Chemistry, University of Warsaw, Warsaw, Poland
- Isabella Felli
- CERM and Department of Chemistry Ugo Schiff University of Florence, Italy
- Witold Andrałojć
- Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland
- Christina Thiele
- Technische Universität Darmstadt, Clemens Schöpf Institut für Organische Chemie und Biochemie, Germany
- Miloš Hricovíni
- Institute of Chemistry, Slovak Academy of Sciences, Bratislava, Slovakia
- Michał Bielejewski
- Institute of Molecular Physics, Polish Academy of Sciences, Poznan, Poland
- Predrag Novak
- Faculty of Science, University of Zagreb, Croatia
- Svetlana Simova
- Bulgarian NMR Centre, Institute of Organic Chemistry with Centre of Phytochemistry Bulgarian Academy of Sciences, Sofia
- Krzysztof Kazimierczuk
- Centre of New Technologies, University of Warsaw, Warsaw, Poland
- Władysław Węglarz
- Institute of Nuclear Physics, Polish Academy of Sciences, Cracow, Poland
- Bruker day
- Eric Leonardis
- Claire Cannet
- Daniel Mathieu
- Frank Schumann
- Oezlen Yasar
- Sylwia Kasprzak
- Pavel Kessler
- Sandra Ulrich
- Christian Steffen
- Klaus Zick
- Sebastian Wegner
- Bruker BioSpin GmbH, Rheinstetten, Germany
- Gerhard Althoff
- Solid state NMR workshop
- Jean-Paul Amoureux
- University of Lille, France
- Sebastian Wegner
- Bruker BioSpin GmbH, Rheinstetten, Germany
- Piotr Paluch
- Faculty of Chemistry, University of Warsaw, Warsaw, Poland
Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences, Lodz, Poland - Alons Lends
- Latvian Institute of Organic Synthesis, Riga, Latvia
- Rasmus Linser
- Technical University Dortmund, Dortmund, Germany
- Zdeněk Tosner
- Charles University in Prague, Prague, Czech Republic
- Dominik Kubicki
- University of Warwick, Warwick, United Kingdom
- Jan Stanek
- Faculty of Chemistry, University of Warsaw, Warsaw, Poland
- Luis Mafra
- CICECO - Aveiro Institute of Materials, Department of Chemistry, University of Aveiro, 3810-193 Aveiro, Portugal
- Marta Dudek
- Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences, Lodz, Poland
- Oliver Pecher
- Department of Chemistry, University of Cambridge, Cambridge, United Kingdom
| Thursday September 22, (Bruker day) |
|
|---|---|
| 9:00 - 9:10 | Welcome and Bruker News Eric Leonardis |
| 09:10 - 09:30 | NMR (at) 1 GHz Daniel Mathieu |
| 09:30 - 09:50 | New probes and Helium recovery Frank Schumann |
| 09:50 - 10:10 | TopSpin News and software tips and tricks Pavel Kessler |
| 10:10 - 11:00 | Coffee break and poster session |
| 11:00 - 11:20 | EPR applications on energy topics Oezlen Yasar |
| 11:20 - 11:40 | News and Updates on Soft- and Hardware Sylwia Kasprzak |
| 11:40 - 12:00 | Introduction to the newly Bruker NMR-based Metabolomics solution for Long COVID Claire Cannet |
| 12:10 - 14:00 | Lunch break |
| 14:00 - 14:20 | Service Updates Sandra Ulrich |
| 14:20 - 14:40 | Fourier 80 - The evolution since 2019 Christian Steffen |
| 14:40 - 15:00 | µImaging and Diffusion applications at SB-Magnets Klaus Zick |
| 15:00 - 15:30 | Coffee break |
| 15:30 - 15:50 | Chemist Suite and other software tools Jerome Coutant |
| 15:50 - 16:10 | Higher sensitivity, better resolution & increased reliability for HR-&CP-MAS Sebastian Wegner |
| 16:10 - 16:30 | Automation in solid state NMR using IconNMR Gerhard Althoff |
| 16:30 - 16:40 | Q&A All |
| 16:40 | Poster price scientific committee |
| 19.00 | Conference dinner |
Lecture abstracts:
Intrinsically disordered Microtubule Associated Protein 2c (MAP2c): Function and regulation probed by NMR
Lukáš Žídek
Faculty of Science and CEITEC, Masaryk University, Brno, Czech Republic
Relaxation properties of intrinsically disordered proteins (IDPs) make NMR investigation of IDPs more sensitive than studies of well-folded proteins. On the other hand, limited dispersion of chemical shifts of IDPs represents a challenge for the resonance assignment. High resolution approaches, typically combining high dimensionality and long evolution times with non-uniform sampling and accelerated return to equilibrium, are needed especially for IDPs of large size and repetitive amino-acid sequences.
Microtubule associated protein 2c (MAP2c) is a 50 kDa IDP controlling microtubule dynamics in the developing brain and playing other, so-far poorly understood, physiological roles. NMR spectroscopy was used to characterize structural features of MAP2c [1,2] and to probe intermolecular interactions and posttranslational modifications important for the MAP2c function and its regulation. In particular, phosphorylation kinetics by Ser/Thr and Tyr kinases and interactions with regulatory proteins have been monitored with resolution of individual amino acids [1,3]. Cross-talks between different signaling pathways have been observed, documenting complexity of regulatory mechanisms controlling MAP2c functions.
This work was supported by Czech Science Foundation, grant number 20-12669S.
[1] Jansen et al., J Biol Chem 292 (2017) 6715
[2] Melková et al., J Biol Chem 293 (2018) 13297
[2] Plucarová et al., J Biol Chem 298 (2022), published online, doi: 10.1016/j.jbc.2022.102384
NMR Spectroscopy in Biomedicine and Structure-Based Drug Design
Robert Konrat
Department of Structural and Computational Biology, Max F. Perutz Laboratories, University
of Vienna, Vienna Biocenter Campus 5, A-1030 Vienna, Austria;
The design of small molecules that bind target proteins with high specificity and affinity is based on the optimization of complementary interactions in the bound state versus the respective unbound solvated states. One crucial interaction in drug design is that between the π-electrons of aromatic ring-systems and aromatic or aliphatic CH containing groups. These so-called CH-π interactions are the most common type of non-covalent interaction in protein-ligand interactions. Here fast and reliable detection methods of bound ligand proton CSPs for the identification and optimization of favorable CH-π interactions between ligand hydrocarbons (CHligand) and protein aromatic amino acids (π protein) are presented.
Intrinsically disordered proteins (IDPs) are challenging the established structural biology structure-function paradigm and thus mandate suitable theoretical framework and concepts to properly address the subtle interdependence between protein structure and dynamics. In particular, it is their structural flexibility that allows them to adapt to and interact with different binding partners, making IDPs suited for functioning as hubs between several interaction partners. In contrast to stably folded globular proteins IDPs sample larger conformational spaces comprising loosely folded as well as compact conformational substates. In order to adequately grasp the subtleties of state distributions of IDPs appropriate theoretical and experimental techniques are needed. Nuclear magnetic resonance (NMR) has matured into a powerful experimental method for the characterization of IDPs and their complexes in solution. In combination with novel computational protein sequence analysis tools it can be used to provide a more comprehensive view of IDPs and how they can simultaneously maintain modularity, robustness, and adaptability, which are essential features for the generation of structural and phenotypic variation in biological systems.
What links NMR with molecular chirality?
Piotr Garbacz
Faculty of Chemistry, University of Warsaw, Poland
Tensors describing nuclear interactions like, e.g., spin-spin coupling, in a chiral molecule and its enantiomer are qualitatively the same except for a usually overlooked antisymmetric part. The unique feature of the antisymmetric part is its behavior under symmetry operations – it is a pseudovector. However, the antisymmetric part considered alone does not indicate the handedness of a molecule. To see chirality directly, i.e., purely on the physical basis, one needs to combine the antisymmetric part of a nuclear tensor with a quantity that transforms like a polar vector, e.g., a permanent electric dipole moment of the molecule. As the molecular electric dipole couples with an electric field, utilizing the antisymmetric part to see chirality requires an extension of nuclear resonance beyond magnetic interaction to magnetoelectric phenomena that may be used for direct discrimination of enantiomers.
Acknowledgments:
PG acknowledges the National Science Centre, Poland, for the financial support through OPUS 16 Grant No. 2018/31/B/ST4/02570.
REFERENCES:
[1] P. Garbacz, Computations of the chirality-sensitive effect induced by an antisymmetric indirect spin-spin coupling, Molecular Physics 116, 1397-1408 (2018)
[2] P. Garbacz, A.D. Buckingham, Chirality-sensitive nuclear magnetic resonance effects induced by indirect spin-spin coupling, Journal of Chemical Physics 145, 204201 (2016)
[3] P. Garbacz, J. Vaara, Direct enantiomeric discrimination through antisymmetric hyperfine coupling, Chemical Communications 57, 8264-8267 (2021)
13C direct detection NMR reveals interesting insights on intrinsically disordered proteins
Isabella C. Felli
CERM and Department of Chemistry Ugo Schiff, University of Florence, Italy
Intrinsically disordered proteins (IDPs) have recently attracted the attention of the scientific community. They sample many different conformations in rapid interconversion, challenging well accepted concepts, such as the structure-function paradigm, stating that a protein functions thanks to its three dimensional structure. When part of complex multi-domain proteins, intrinsically disordered regions (IDRs) often play a key role in modulating the properties of the protein itself. However they pose serious challenges to most of the currently used methods to achieve atomic level information. Current drug discovery approaches based on seeking the optimal fit between complementary well defined surfaces (lock-and-key paradigm) are also bound to fail if applied to IDPs/IDRs.
NMR spectroscopy represents an excellent tool (if not the unique one) to access atom-resolved information about the structural and dynamic properties of highly flexible IDPs/IDRs. NMR methods should be optimized to overcome critical points deriving from the absence of the 3D structure. The impact on NMR observables deriving from the peculiar structural and dynamic properties of IDPs/IDRs will be discussed highlighting how 13C direct detection contributes to overcoming critical points.
Several examples of particular features or motives that often occur in IDPs/IDRs will be presented, revealing novel structural and dynamic modules that are not yet described in the PDB.
Investigating Thermoresponsive Alignment Media via 2H NMR and Diffusion
Christina M. Thiele, Dominic S. Schirra, Timo Imhof, Jonas Kind
Technische Universität Darmstadt, Darmstadt, Germany
cthiele(at)thielelab.de
Investigating an anisotropic NMR parameter become increasingly important in organic structure elucidation for the determination of conformation and relative configuration of natural products, synthesized compounds and catalysts.[1] To obtain such anisotropic NMR parameter, suitable alignment media are necessary. The use of lyotropic liquid crystals based on helically chiral polymers is especially intriguing as they additionally allow for enantiodiscrimination.[2] We have recently synthesized several homopolypeptides, which form lyotropic liquid crystals that are suitable for the measurement of anisotropic NMR observables and show excellent enantiodiscrimination. Furthermore, they exhibit thermoresponsive properties.[3]
The intriguing properties of these new thermoresponsive alignment media will be described in this presentation. Furthermore, we will shed light on the processes responsible for the thermoresponsivity by using isotope labelling, 2H NMR[4] and anisotropic diffusion[5].
REFERENCES
[1] For reviews see: C. M. Thiele, Eur. J. Org. Chem. 2008, 5673-5685; V. Schmidts, Magn. Reson. Chem. 2017, 55, 54-60.
[2] For review see: P. Lesot, J. Courtieu, Prog. Nucl. Magn. Reson. Spectrosc. 2009, 55, 128-159.
[3] M. Schwab, D. Herold, C. M. Thiele, Chem. Eur. J. 2017, 23, 14576-14584; M. Schwab, V. Schmidts, C. M. Thiele, Chem. Eur. J. 2018, 24, 14373-14377; S. Jeziorowski, C. M. Thiele, Chem. Eur. J. 2018, 24, 15631-15637.
[4] D. S. Schirra, M. Hirschmann, I. A. Radulov, M. Lehmann, C. M. Thiele, Angew. Chem. 2021, 133, 21208-21214.
[5] T. Imhof, J. Kind, C. M. Thiele, manuscript in preparation.
NMR and DFT analysis of 3D structure and spin-spin coupling constants in biologically active saccharides
Miloš Hricovíni
Institute of Chemistry, Slovak Academy of Sciences, 845 38 Bratislava, Slovakia
Milos.Hricovini(at)savba.sk
Many oligo- and polysaccharides participate in various important biological processes and the molecular basis of these processes has been subject of a number of NMR and theoretical studies. Highly negatively charged glycosaminoglycans - heparin and heparan sulphate belong to the most studied saccharides. Apart from high-resolution NMR data, theoretical analysis is an important part of our understanding of biological properties of heparin and heparan sulphate oligosaccharides. In the present contribution, the results of NMR analysis and density functional theory (DFT) calculations are discussed. Geometry optimizations, evaluating explicit solvent molecules, were performed by the B3LYP/6311+G(d,p) methods. The optimized molecular geometries in various heparin oligosaccharides allowed calculations of NMR parameters, such as chemical shifts and coupling constants. Comparison of theoretical and experimental NMR data showed that DFT method using the B3LYP functional and the 6311+G(d,p) basis set, combined with explicit solvent model, can yield sufficiently accurate structural and NMR data for these oligosaccharides.
Dynamics and proton transport in noncrystalline cellulose functionalized with imidazolium cations
Michał Bielejewski, Jadwiga Tritt-Goc
Institute of Molecular Physics Polish Academy of Sciences, Poznań, Poland
michal.bielejewski(at)ifmpan.poznan.pl
Proton conductors are one of the key elements of modern power sources like fuel cells which converts the chemical energy of hydrogen and oxygen reaction into electricity cleanly and efficiently. To overcome technical limitations and to create durable, cost-efficient, high-performance, and environmentally neutral materials for membranes used in FCs, we have designed and synthesized a nanocomposite based on cellulosic materials and heterocyclic molecules. The nanocrystalline cellulose was used to prepare the foil functionalized with imidazole molecules via a modified sol-gel technique to obtain a new solid proton conductor [1,2]. The 1 H, 15N, and 13C CP-MAS NMR spectroscopy has been used to study the dynamical properties of the cation molecule to determine the mechanism and efficiency of protonic transport in studied membranes [3,2]. This presentation will present the possibility of using 13C NMR spectroscopy as an alternative to 15N NMR spectroscopy.
REFERENCES
[1] M. Bielejewski, Ł. Lindner, R. Pankiewicz, J. Tritt-Goc Cellulose 2020, 27, 1989–2001.
[2] Ł. Lindner, M. Bielejewski, E. Markiewicz, A. Łapiński, R. Pankiewicz, J. Tritt-Goc Eur. Polym. J. 2021, 161, 110825.
[3] J. Tritt-Goc, Ł. Lindner, M. Bielejewski, E. Markiewicz, R. Pankiewicz Carbohydr. Polym. 2019, 225, 115196
[4] M. Bielejewski, M. Pinto-Salazar, Ł. Lindner, R. Pankiewicz, G. Buntkowsky, J. Tritt-Goc J. Phys. Chem. C. 2020, 124, 18886-18893
[5] J. Tritt-Goc, Ł. Lindner, M. Bielejewski, E. Markiewicz, R. Pankiewicz Int. J. Hydrog. Energy 2020, 45, 13365-13375
Solution structure of a lanthanide-binding DNA aptamer determined using high quality pseudocontact shift restraints
Witold Andrałojć, Julia Wieruszewska, Karol Pasternak, Zofia Gdaniec
Institute of Bioorganic Chemistry, Polish Academy of Sciences, Noskowskiego 12/14, 61-704 Poznan, Poland
wandralojc(at)ibch.poznan.pl
NMR spectroscopy is among the primary techniques used for high resolution structural studies of nucleic acid systems. Classical approaches to biomolecular NMR structure determination rely of the measurement of inter-proton distances up to 5-6 Å, through the means of the Nuclear Overhauser Effect (NOE). The local nature of these distance restraints can lead to a poor reproduction of some global structural features of the studied molecule (e. g. the degree of helical bending). Long-range NMR structural restraints derived from the presence of a paramagnetic ion in the studied system can radically change this situation and have already found much success in NMR studies of proteins, beyond just structure determination1,2. However, the potential of paramagnetic NMR methods remains largely unrealized for nucleic acids due to lack of general methods to rigidly and site-specifically introduce paramagnetic ions into these systems.
Here we present the high-resolution NMR structure of a lanthanide-binding DNA aptamer and demonstrate that lanthanide ions bound to this system induce large magnitude paramagnetic effects (pseudocontact shifts; PCS), readily interpretable in terms of structural parameters. We propose that the lanthanide binding motif uncovered for this system can be exploited to introduce lanthanide binding sites into a broad range of different DNA and RNA molecules to facilitate their structural and functional NMR studies. The presented structure is also of interest by itself, as it constitutes the first high-resolution structure of a metal-binding aptamer deposited in the PDB, furthering our understanding of high-affinity metal binding by DNA.
Acknowledgment: This work has been supported by the National Science Center (Poland) under grant agreements: [UMO-2020/37/B/ST4/03182] and [UMO-2018/31/D/ST4/01467] to W.A.. The calculations were performed at Poznań Supercomputing and Networking Center.
REFERENCES
[1] G. Otting, “Protein NMR using paramagnetic ions.”, Annu. Rev. Biophys., 2010, 39, 387-405.
[2] C. Luchinat, G. Parigi, E. Ravera, “Paramagnetism in Experimental Biomolecular NMR.” (The Royal Society of Chemistry, 2018), ISBN 978-1-78801-086-3
Exploring macrolide binding modes by NMR spectroscopy
Predrag Novak
University of Zagreb, Faculty of Science, Department of Chemistry, Zagreb, Croatia
15-membered macrolides are well-known class of antibacterials widely prescribed to treat upper and lower respiratory tract infections [1]. Azithromycin is a semi-synthetic macrolide antibiotic derived from erythromycin possessing broad spectrum of antibacterial potency and favorable pharmacokinetics. Similar to other macrolide antibiotics it exerts its activity by binding to the bacterial ribosomal 23S rRNA in domain V at the peptidyl transferase region and blocks the exit tunnel which in turn inhibits the bacterial protein synthesis. However, bacterial resistance to marketed antibiotics is growing rapidly and represents one of the major hazards to human health worldwide. Recently discovered conjugates of azithromycin and thiosemicarbazones, the macrozones, represent new class of macrolide derivatives that exhibit promising activities against resistant pathogens [2-3].
Interaction studies of macrolide antibiotics and some selected macrozones with their biological targets will be presented in this talk. NMR experiments such as transferred nuclear Overhauser effect spectroscopy (trNOESY), saturation transfer difference (STD), diffusion and solvent paramagnetic relaxation experiments (PRE) in combination with molecular modelling have been employed to characterize binding modes and epitopes and asses bound conformations [4-6].
These data can serve as a platform for design of novel inhibitors with an improved biological profile.
Acknowledgments:
This work has been financially supported by the Croatian Science Foundation (project Macrozones, IP-2018-01-8098).
REFERENCES:
[1] B. Arsic, P. Novak, M.G. Rimoli, J. Barber, G. Kragol, F. Sodano, Macrolides. Properties, Synthesys and Applications. Berlin: De Gruyter; 2018.
[2] I. Grgičević, I. Mikulandra, M. Bukvić, M. Banjanac, I. Habinovec, B. Bertoša, P. Novak, Int. J. Antimicrob. Agents 2020, 56, 106147.
[3] I. Mikulandra, T. Jednačak, B. Bertoša, J. Parlov-Vuković, I. Kušec and P. Novak, Materials 2021, 14, 5561
[4] S. Glanzer, S. A. Pulido, S. Tutz, G.E. Wagner, M. Kriechbaum, N. Gubensäk, J. Trifunovic, M. Dorn, W. M. F. Fabian, P. Novak, J. Reidl, K. Zangger, Chem. Eur. J. 2015, 21 4350.
[5] S. Kosol, E. Schrank, M. Bukvić Krajačić, G. E. Wagner, H. Meyer, C. Göbl, G.N. Rechberger, K. Zangger, P. Novak, J. Med. Chem. 2012, 55, 5632.
[6] P. Novak, J. Barber, A. Čikoš, B. Arsić, J. Plavec, G. Lazarevski, P. Tepeš and N. Košutić-Hulita, Bioorg. Med. Chem. 2009, 17, 5857.
Do we know what we eat - food metabolomics?
Dessislava Gerginova1, Biljana Smit2, Svetlana Simova1
1Institute of Organic Chemistry with Centre of Phytochemistry, Acad. G. Bonchev bl. 9, 1113 Sofia, Bulgaria
2Department of Science, Institute for Information Technologies, University of Kragujevac, Jovana Cvijića bb, 34000 Kragujevac, Serbia
Metabolomics aims to profile all chemical compounds present in a system. Numerous chemicals can be monitored simultaneously by using NMR or MS without prior knowledge of which compounds are altered. Investigations in food science include how a food product changes with food processing, to detect adulteration or misbranding or changes in nutritional profile, to understand what flavor compounds contribute to product liking, and to explore how dietary interventions with these food products alter the human metabolome. Data can also be correlated with other information, e.g. sensory evaluation data, genetic analyses or microbiological data to provide additional value.
On the example of a set of samples collected from a certain area a series of problems will be demonstrated indicating deficiencies in the information we get about the origin and composition of an everyday used food product.
Acknowledgements: This work was supported by the National Research Program “Healthy Foods for a Strong Bio-economy and Quality of Life” using the distributed research infrastructure INFRAMAT, Ministry of Education and Science.
REFERENCES
[[1] E. Hatzakis (2019) Comprehensive Reviews in Food Science and Food Safety, 18, 189-220;
[2] O. Adebo, P. Njobeh, J. Adebiyi, S. Gbashi & E. Kayitesi (2017) Food Metabolomics: doi: 10.5772/intechopen.69171;
[3] J. Selamat, N. Rozani, S. Murugesu, S. (2021) Application of the Metabolomics Approach in Food Authentication. Molecules, doi: 10.3390/molecules26247565.
Methods for exploring non-Fourier dimensions - from small molecules to proteins.
Krzysztof Kazimierczuk
Centre of New Technologies, University of Warsaw, Poland
The possibility of adding extra dimensions to spectra made the NMR spectroscopy a powerful analytical tool. Besides commonly used indirect „Fourier dimensions”, i.e., those processed with Fourier transform, we often acquire a series of spectra under varying experimental conditions. These can be pulse sequence parameters (e.g. mixing times or diffusion-encoding gradients), environmental conditions (temperature, pH or concentration) or reaction progress.[1]
The resulting stack of n-dimensional spectra may be considered as an n+1-dimensional object. This concept opens the way to new processing methods for exploring non-Fourier dimensions.
Over the last decade, my group has developed several methods based on time-resolved non-uniform sampling (TR-NUS), Radon transform, and non-stationary signal processing. In my presentation, I will recapitulate the theory of these approaches and show their applications in metabolomics[2], reaction monitoring[3], ligand binding[4] and studying protein dynamics[5]. I will also discuss the currently developed concepts of exploring non-Fourier dimensions in protein research, e.g., fast measurements of TOCSY transfer curves.
REFERENCES
[1] Gołowicz, D. et al. (2020). Fast time-resolved NMR with non-uniform sampling. Progress in Nuclear Magnetic Resonance Spectroscopy, 116, 40–55.
[2] Nawrocka, E. K. et al. (2021). Variable-temperature NMR spectroscopy for metabolite identification in biological materials. RSC Advances, 11(56), 35321–35325.
[3] Nawrocka, E. K. et al. (2019). Nonstationary Two-Dimensional Nuclear Magnetic Resonance: A Method for Studying Reaction Mechanisms in Situ. Analytical Chemistry, 91(17), 11306–11315.
[4] Romero, J. A. et al. (2020). Non-Stationary Complementary Non-Uniform Sampling (NOSCO NUS) for Fast Acquisition of Serial 2D NMR Titration Data. Angewandte Chemie - International Edition, 59(52), 23496–23499.
[5] Shchukina, A. et al. (2021). Temperature as an Extra Dimension in Multidimensional Protein NMR Spectroscopy. Chemistry – A European Journal, 27(5), 1753–1767.
Magnetic Resonance Imaging in Biomedical, Pharmaceutical and Geological Sciences.
Władysław P. Węglarz
Institute of Nuclear Physics Polish Academy of Sciences, Kraków, Poland
During the last four decades the Magnetic Resonance Imaging (MRI) and Spectroscopy (MRS) become a very powerful diagnostic modality, due to possibility of non-invasive visualisation of the human body. However, besides of the clinical applications, it is also used as a useful research tool in preclinical bio-medicine, for testing new pharmacological or theranostic therapies using animal models of the civilisation diseases and imaging of the structure, functions and metabolisms of the living organisms. It is also used in different branches of material sciences for e.g. characterisation of the properties of new theranostics and contrast agents as well as pharmaceutical, food or porous materials. Such broad range of applications is available due to possibility of obtaining images and localised NMR spectra, dependent on different physico-chemical properties, including nuclear relaxation times, diffusion, perfusion, magnetisation exchange, or presence of specific metabolites, to name a few. In most cases the proton (1H) MRI is utilized, however other nuclei, like 19F, 31P, 23Na or 39K are investigated as well. In the present lecture a few examples of using different MRI/MRS techniques for investigations of the brain structure and metabolism using animal models in vivo and ex vivo[1-3], optimising contrasting properties of the novel theranostic agents for drug delivery[4-6], testing novel pharmaceutical dosage forms properties and in vivo distribution[7] and finally for characterisation of the oil- and gas-bearing porous rocks[8-9] will be presented.
REFERENCES
[1] P. Majka, N. Chłodzińska, K. Turlejski, T. Banasik, R.L. Djavadian, W.P. Węglarz, D.K. Wójcik, A three-dimensional stereotaxic atlas of the gray short-tailed opossum (Monodelphis domestica) brain, Brain. Struct. Funct., 223 (2018) 1779-1795
[2] K. Gzieło, K. Janeczko, W. Węglarz, K. Jasiński, K. Kłodowski, Z. Setkowicz, MRI spectroscopic and tractography studies indicate consequences of long-term ketogenic diet, Brain. Struct. Funct., 225 (2020) 2077-2089
[3] K. Skowron, K. Jasiński, W.P. Węglarz et al., Is the Activity-Based Anorexia Model a Reliable Method of Presenting Peripheral Clinical Features of Anorexia Nervosa?, Nutrients, 13 (2021) 2876
[4] A. Maximenko, J. Depciuch, N. Łopuszyńska, Ż. Świątkowska-Warkocka, P.M. Zieliński, W.P. Węglarz, M. Parlińska-Wojtan et al., Fe3O4(at)SiO2(at)Au nanoparticles for MRI-guided chemo/NIR photothermal therapy of cancer cells, RSC Adv., 10 (2020) 26508-20
[5] M. Szczęch, D. Orsi, N. Łopuszyńska, L. Cristofolini, K. Jasiński, W.P. Węglarz, F. Albertini, S. Kereiche, K. Szczepanowicz, Magnetically responsive polycaprolactone nanocarriers for application in the biomedical field: magnetic hyperthermia, magnetic resonance imaging, and magnetic drug delivery, RSC Adv., 71 (2020) 43607-18
[6] N. Łopuszyńska, K. Szczepanowicz, K. Jasiński, P. Warszyński, W.P. Węglarz, Effective Detection of Nafion®-Based Theranostic Nanocapsules through 19F Ultra-Short Echo Time MRI, Nanomaterials, 10 (2020) 2127
[7] G. Wyszogrodzka-Gaweł, P. Dorożyński, S. Giovagnoli, W. Strzempek, E. Pesta, W.P. Węglarz, B. Gil, E. Menaszek, P. Kulinowski, An Inhalable Theranostic System for Local Tuberculosis Treatment Containing an Isoniazid Loaded Metal Organic Framework Fe-MIL-101-NH2—From Raw MOF to Drug Delivery System, Pharmaceutics, 11 (2019) 687
[8] W.P. Węglarz, A. Krzyżak, G. Machowski, M. Stefaniuk, ZTE MRI in high magnetic field as a time effective 3D imaging technique for monitoring water ingress in porous rocks at sub-millimetre resolution, Magn. Reson. Imaging, 47 (2018) 54-59
[9] A.T. Krzyżak, W. Mazur, A. Fheed, W.P. Węglarz, Prospects and Challenges for the Spatial Quantification of the Diffusion of Fluids Containing 1H in the Pore System of Rock Cores, J. Geophys. Res. Solid Earth, 127 (2022) 1-20
Recent developments in NMR of quadrupolar nuclei in solids
Trébosc,1 Lafon1, Hung2, Gan2, Nagashima3, Sasaki4, Bayzou1, Amoureux1,5
1Lille-France, 2Tallahassee-US, 3Tsukuba-Japan, 4Bruker-Japan, 5Bruker-France
Over 74% of NMR active nuclei have a spin I ≥ 1 and are subject to quadrupole interactions. Nevertheless, the observation of these nuclei in solids often remains more challenging than with spin-1/2 nuclei since: (i) the spectral resolution is decreased by the second-order quadrupolar interaction, (ii) the larger size of the density matrix complicates the spin dynamics, and (iii) the quadrupole interactions exceed the rf-field amplitude. We will present recent techniques we introduced to facilitate the observation of these quadrupolar isotopes.
We have analyzed the performances of the recent cosine low-power MQMAS sequence,[1] and showed that for spin-3/2 isotope, this technique is more efficient than STMAS and requires lower rf-field; whereas for spin-5/2 nuclei, this variant is as efficient as the high-power one but requires rf-fields smaller than 20 kHz and hence, can be employed for low-γ nuclei and large diameter rotors.[2] These advantages are obtained without the need for the so-called STMAS specifications, and the sequences have been extended to obtain MQ-HETCOR spectra.
We have also developed efficient pulse sequences to transfer magnetization from protons to quadrupolar nuclei at slow-moderate or fast MAS (νR = 10-25 or > 60 kHz).[3-5] For νR ≤ 20 kHz, the most efficient pulse sequence is D-RINEPT using adiabatic pulses. This transfer has been combined with DNP to detect low-γ quadrupolar nuclei with natural abundance near surfaces or sub-surfaces, such as 17O, 95Mo, 47,49Ti, 67Zn.[5] More recently, this transfer has been combined with MQMAS to detect DNP-enhanced high-resolution NMR spectra of quadrupolar nuclei, such as 17O, near surfaces (Fig).[6]
Finally, we have analyzed the T-HMQC technique[7] for the indirect detection without t1 noise of spin-1/2 nuclei subject to large CSA (195Pt), as well as spin-1 (14N) and spin-3/2 (35Cl) quadrupolar nuclei.[8] For spin-3/2 nuclei, this method can provide a resolution enhancement of ca. 4.[9]
Acknowledgment: This work has been supported by the National Science Center (Poland) under grant agreements: [UMO-2020/37/B/ST4/03182] and [UMO-2018/31/D/ST4/01467] to W.A.. The calculations were performed at Poznań Supercomputing and Networking Center.
REFERENCES
[1] I. Hung, Z. Gan, JMR, 324, 106913, 2021; Z. Gan, I. Hung, JMR, 328, 106994, 2021.
[2] A. Sasaki, et al, submitted.
[3] J. S. Gomez, et al, MR, 2, 447, 2021.
[4] H. Nagashima, et al, MRC, 1-20, 2021.
[5] H. Nagashima, et al, JACS, 142, 10659, 2020.
[6] H. Nagashima, et al, submitted.
[7] I. Hung, P. Gor’kov, Z. Gan, JCP, 151, 154202, 2019; I. Hung, Z. Gan, JPCL, 11, 4734, 2020.
[8] R. Bayzou, J. Trébosc, I. Hung, Z. Gan, O. Lafon, JP. Amoureux, JCP, 2022.
[9] R. Bayzou, J. Trébosc, Z. Gan, I. Hung, O. Lafon, JP. Amoureux, submitted.
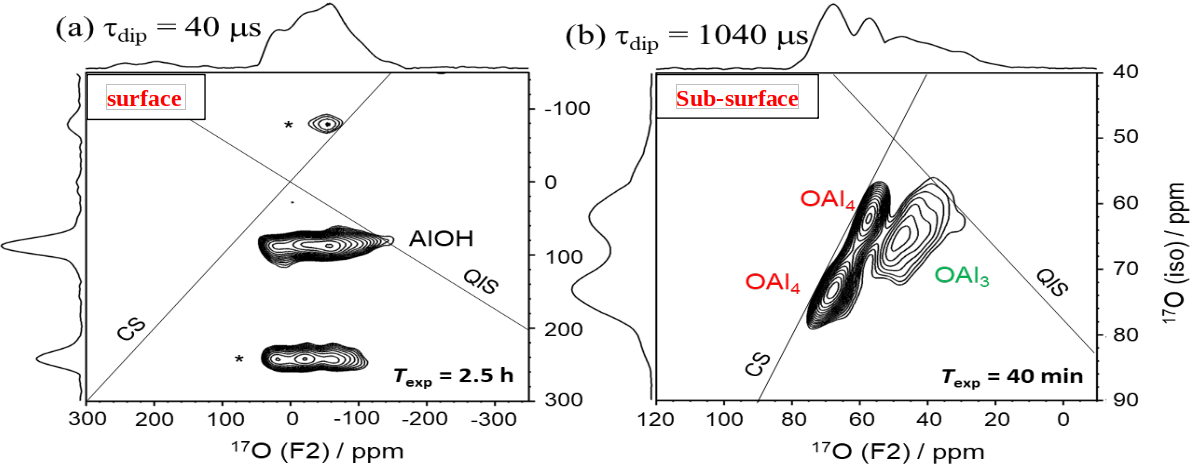
2D DNP-enhanced 1H →17O D-RINEPT-MQMAS spectra of γ-Al2O3 with Τdip = (a) 40 or (b) 1040 μs
New salts of teriflunomide (TFM) – Single crystal X-ray and solid state NMR investigation
Piotr Paluch1,2, Tomasz Pawlak1, Rafał Dolot1, Grzegorz Bujacz3, Marek J.Potrzebowski1
1Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences
2Faculty of Chemistry, Biological and Chemical Research Centre, University of Warsaw
3Institute of Molecular and Industrial Biotechnology, Lodz University of Technology
New salts of teriflunomide TFM (drug approved for Multiple Sclerosis treatment) with inorganic counterions: lithium (TFM_Li), sodium (TFM_Na), potassium (TFM_K), rubidium (TFM_Rb), caesium (TFM_Cs) and ammonium (TFM_NH4) were prepared and investigated employing solid state NMR Spectroscopy, Powder X-ray Dif- fraction PXRD and Single Crystal X-ray Diffraction (SC XRD). Crystal and molecular structures of three salts: TFM_Na TFM_Cs and TFM_NH4 were determined. Compared to the native TFM, for all crystalline salt structures, a conformational change of the teriflunomide molecule involving about 180-degree rotation of the end group, forming an intramolecular hydrogen bond N–H⋯O is observed. By applying a complementary multi-technique approach, employing 1D and 2D solid state multinuclear (1H,13C, 15N, 19F, 23Na, 7Li, 133Cs) MAS NMR techniques, single and powder X-ray diffraction measurements, as well as the DFT-based GIPAW calculations of NMR chemical shifts for TFM_Na and TFM_Cs allowed to propose structural features of TFM_Li for which it was not possible to obtain adequate material for single crystal X-Ray measurments.
Acknowledgments:
The computational resources were partially provided by the Polish Infrastructure for Supporting Computational Science in the European Research Space (PL-GRID). The project no. 04.04.00-00-4374/17-01 (Homing/2017 4/37) is carried out within the HOMING programme of the Foundation for Polish Science co-financed by the European Union under the European Regional Development Fund
REFERENCES:
Tomasz Pawlak, Piotr Paluch, Rafał Dolot, Grzegorz Bujacz, Marek J.Potrzebowski, Solid State NMR 10.1016/j.ssnmr.2022.101820
Amyloid fibrils and intact cells seen by multinuclear solid-state NMR spectroscopy at fast MAS regime
Alons Lends
Latvian Institute of Organic Synthesis, Riga
The solid-state NMR (ssNMR) spectroscopy is a powerful technique to study biological systems at atomic level close to their native state. Recently fast MAS techniques has revolutionized ssNMR spectroscopy, by requiring less 1 mg of sample material. Here we are going to present the application of ssNMR to study amyloid fibrils and intact cells. In the first part of presentation, we are going to explore 19F detection methods with a novel fluorine labelling methods to study amyloid fibrils at fast MAS regime. We manage to achieve an excellent resolution in 19F dimension. This technique will be further explored to obtain unambiguous distance restraints for the 3D structure calculations. Additionally, we will also show the incorporation of selectively 2H labelled amino acids into proteins to apply 2H detection methods for the specific dynamic measurements. In the second part of the talk, we will demonstrate the 1H detection methods to study intact yeast cells. For this sample we manage to achieve a moderate resolution with a good sensitivity. This allowed us to identify the main structural components and dynamics of the cells.
Conformational exchange and proton dynamics – from DNA recognition to enzymatic catalysis by NMR
Rasmus Linser
Faculty of Chemistry, Biological and Chemical Research Centre, University of Warsaw
Conformational exchange and proton dynamics – from DNA recognition to enzymatic catalysis by NMR
Structural dynamics are the fundament of biomolecular functionality. NMR gives atomic-resolution insights for conformational exchange as well as fluctuations of chemical constitution at an unsurpassed level of detail. Here I will give an overview over some of the current questions we are tackling in our group, reaching from a dynamics-optimized artificial epigenetic reader to micro-crystalline enzymes, touching on the virtues of fast magic-angle spinning and higher-dimensionality, proton-detected experiments.
Improving sensitivity of solid-state NMR of proteins by optimal control methods
Zdeněk Tošner
Charles University in Prague, Prague, Czech Republic
We have recently developed several strategies to improve sensitivity of solid-state NMR experiments of protein samples. Traditional cross-polarization methods used to establish inter-nuclear correlations suffer from radiofrequency field inhomogeneity and lead to volume selection when only part of a sample yields NMR signal. Our tm-SPICE optimal control pulses are designed to compensate for this inhomogeneity and yield sensitivity gains of about 50% for the 15N-13C transfer. A new concept of TROP pulses explores the preservation of equivalent pathways principle in multidimensional experiments known from liquid state NMR. Transferring both x- and y- magnetization components after indirect chemical shift evolution leads to the additional 1.41-fold increase of sensitivity per indirect dimension. This provides an order of magnitude time savings in just-emerging 4-5D proton-detected experiments.
New approaches to determining the atomic-level structure of advanced materials
Dominik J. Kubicki
Department of Physics, University of Warwick
@DominikJKubicki(Tweeter), http://kubickilab.wordpress.com/
Determining the structure-property relationships at multiple length scales is one of the key tenets of rational design of new materials. While diffraction techniques offer insight into the long-range structure of solids, many properties are determined by local structure, which can be accessed using approaches based on, e.g., total scattering (PDF), XAFS, and magnetic resonance (NMR and ESR).
I will use the example of metal halide perovskites to discuss how we can determine the atomic-level structure of solids in an element-specific manner using solid-state NMR spectroscopy. The range of research problems includes quantifying dopant incorporation, phase segregation, decomposition pathways, passivation mechanisms, and structural dynamics.[1] I will also show how electron diffraction allows us to study structural phenomena inaccessible with X-rays.[2]
I will then discuss my take on studying these multifaceted materials in situ and operando to elucidate the mechanism of structural transformations in fully assembled optoelectronic devices, especially under illumination. These strategies will be key to elucidating the performance-limiting factors in devices such as solar cells, light emitting diodes, and X-ray detectors.
REFERENCES:
[1] Kubicki, D. J.; Stranks, S. D.; Grey, C. P.; Emsley, L. NMR Spectroscopy Probes Microstructure, Dynamics and Doping of Metal Halide Perovskites. Nat. Rev. Chem. 2021, 5, 624–645.
[2] Doherty, T.; Nagane, S.; Kubicki, D. J.; Jung, Y. K.; Johnstone, D. N.; Iqbal, A.; Guo, D.; Frohna, K.; Danaie, M.; Tennyson, E. M.; MacPherson, S.; Abfalterer, A.; Anaya, M.; Chiang, Y.-H.; Crout, P.; Ruggeri, F. S.; Collins, S.; Grey, C. P.; Walsh, A.; Midgley, P.; Stranks, S. D. Stabilized Tilted-Octahedra Halide Perovskites Inhibit Local Formation of Performance-Limiting Phases. Science 2021, 374, 1598–1605.
NMR assignment of methyl groups using 13C homonuclear transfers, tailored isotope labelling and proton detection with MAS up to 100 kHz
Jan Stanek1, Piotr Paluch1,2, Rafał Augustyniak1, Mai-Liis Org3, Kalju Vanatalu3, Ats Kaldma3, Ago Samoson3
1Faculty of Chemistry, Biological and Chemical Research Centre, University of Warsaw
2Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences
3Tallin University of Technology, Tallinn, Estonia
In NMR spectroscopy of proteins, methyl protons play a particular role as sensitive reporters on protein dynamics, allosteric effects and protein-protein interactions, accessible in solution in high-molecular weight systems approaching 1 MDa. However, their systematic resonance assignment presents a challenge, and often requires a labor-intensive mutagenesis approach. Here we address the issue by using solid-state NMR methods, and leverage recent technological advancements in 1H-detection with magic-angle spinning at frequencies up to and beyond 100 kHz [1].
We employ specific 1H-labelling of Ile, Leu, and Val residues, and otherwise an extensive deuteration and uniform 13C-enrichment, to establish correlations between methyl and backbone amide 1H resonances. To this end, we systematically evaluate efficiency of a series of RF schemes which transfer 13C coherence across entire 13C side-chains of methyl-containing residues. Performance of ten methods for recoupling of either isotropic 13C-13C scalar or anisotropic dipolar interactions (five variants of TOBSY, FLOPSY, DIPSI, WALTZ, RFDR and DREAM) is evaluated experimentally at two magic-angle spinning (55 and 94.5 kHz) and magnetic field conditions (18.8 and 23.5 T).
Model isotopically-labelled compounds and alpha-spectrin SH3 protein are used as convenient reference systems. Results indicate a pivotal role of increased MAS rates for efficiency of multiple-bond 13C transfer even in highly deuterated protein samples. Additionally, spin dynamics simulations in SIMPSON are performed to determine optimal parameters of these RF schemes, up to recently experimentally attained spinning frequencies (200 kHz) and B0 field strengths (30.5 T), and beyond.
We revisit the concept of linearization of 13C side-chains by tailored isotope labelling and show a dramatic sensitivity gain of methyl-to-backbone correlations. We also demonstrate that high-resolution 4D spectroscopy with non-uniform sampling is key to remove ambiguities in simultaneous resonance assignment of methyl proton and carbon chemical shifts. We estimate that a combination of suitable isotope labelling, sensitive 1H-detection, and optimal RF designs for ultra-fast MAS enables methyl resonance assignment in microcrystalline proteins as large as 500 residues [2].
We additionally present the surprising advantages of 2H decoupling for resolution and coherence lifetime of 13C spins in extensively deuterated proteins. In this respect, we revisit the conventional triple-correlation approach (HN-CA/CB/CO) and present ultimate RF designs for backbone resonance assignment with 3D to 5D spectroscopy [1,3], along with recent novel approaches for resonance assignment in nondeuterated proteins [4].
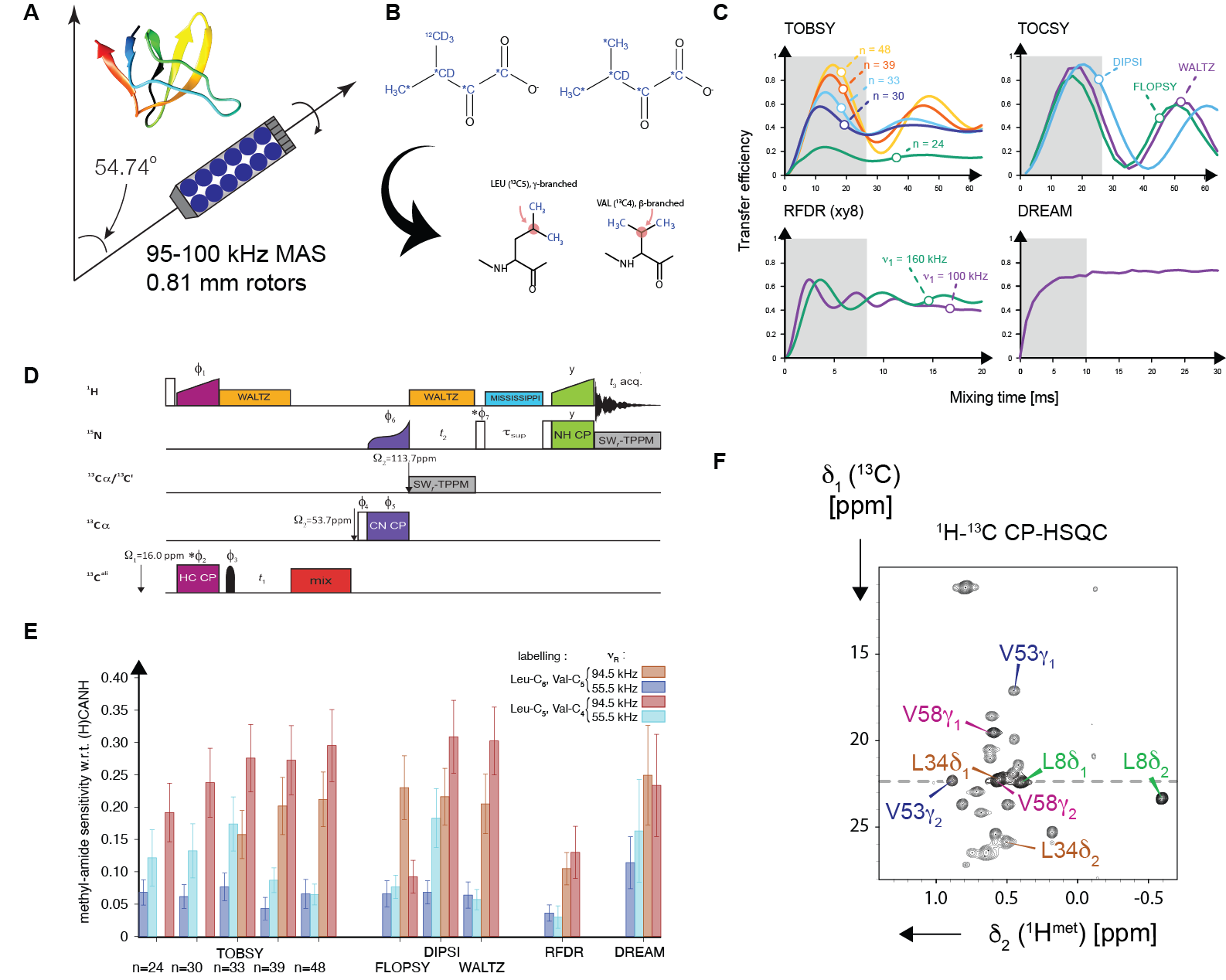
Figure: A combination of (A) ultrafast (95-100 kHz) magic-angle spinning in small (0.81 mm) rotors, (B) suitable precursors for biochemical synthesis of ILV aminoacids, (C) spin dynamics simulations, (D) 3-4D spectroscopy and a careful choice between J- and D-mediated 13C-13C mixing schemes is used in this work for efficient assignment of 1H and 13C chemical shifts of methyl groups (E).
Acknowledgments:
This research was funded by Polish National Science Centre under a grant contract no. 2019/33/B/ST4/02021 and by Polish National Agency for Academic Exchange with generous support of Polish Return programme (Contract No. PPN/PPO/2018/1/00098). The computational resources for this work were provided by the Polish Infrastructure for Supporting Computational Science in the European Research Space (PLGRID, grant ID: plgnmrsi2, plgnmrsi3).
REFERENCES:
[1] Le Marchand T, Schubeis T, Bonaccorsi M, Paluch P, Lalli D, Pell AJ, Andreas LB, Jaudzems K, Stanek J, Pintacuda G, 1H‑Detected Biomolecular NMR under Fast Magic-Angle Spinning, Chem Rev., 122, 9943-10018, 2022
[2] Paluch P, Augustyniak R, Org M-L, Vanatalu K, Kaldma A, Samoson A and Stanek J, NMR Assignment of Methyl Groups in Immobilized Proteins Using Multiple-Bond 13C Homonuclear Transfers, Proton Detection, and Very Fast MAS, Front. Mol. Biosci., 2022, 9:828785, doi 10.3389/fmolb.2022.828785.
[3] Orton HW, Stanek J, Schubeis T, Foucaudeau D, Ollier C, Draney AW, Le Marchand T, Cala-De Paepe D, Felli IC, Pierattelli R, Hiller S, Bermel W, Pintacuda G. Protein NMR Resonance Assignment without Spectral Analysis: 5D SOlid-State Automated Projection spectroscopY (SO-APSY). Angewandte Chemie International Edition, 59(6):2380–2384, 2020
[4]Stanek J, Schubeis T, Paluch P, Guentert P, Andreas LB, Pintacuda G. Automated backbone NMR resonance assignment of large proteins using redundant linking from a single simultaneous acquisition. J. Am. Chem. Soc., 142(12):5793–5799, 2020
Probing gas species adsorbed at the surfaces of porous materials using ssNMR and modeling
Luís Mafra
CICECO - Aveiro Institute of Materials, Department of Chemistry, University of Aveiro, 3810-193 Aveiro, Portugal
This presentation highlights the most recent advances from our team in studying the speciation of gas adsorbates (mainly CO2) and trimethylphosphine oxide (TMPO) probe molecules that interact with the surfaces of porous materials. The nature of adsorbate species formed in confined spaces, their location inside the pore structure and the type of adsorbate-adsorbent interactions, determines many properties in catalytic and CO2-adsorbent materials such as gas sorption capacity/kinetics, selectivity, acid strength and cyclic stability. However, an atomic-level understanding of the adsorbate sorption mechanisms in porous media and the interactions involved therein remains elusive, hindering our ability to design improved catalytic and sorbent materials. The lack of advanced spectroscopic studies, tailored to elucidate the structure of adsorbed gas species, has also been a major bottleneck for further progresses in understanding the physical chemistry of gas-solid interfaces. This talk shows examples on the use of solid-state (ss) NMR spectroscopy has a unique site-selective technique to study the structure and the dynamics of CO2 species adsorbed at porous adsorbent materials. Unprecedented atomistic description of the host–guest and guest–guest interactions of TMPO molecules confined within HZSM-5 molecular-sized voids combining ssNMR, DFT and ab initio molecular dynamics (AIMD)-based computational modeling will also be showcased.
NMR crystallography – demanding but gratifying
Marta K. Dudek
Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences, Sienkiewicza 112, 90-363, Lodz, Poland
NMR crystallography is an excellent approach to describe crystal structures of molecules crystallizing in the form of microcrystalline powders. Typically, it consists of advanced solid-state NMR and powder X-Ray diffraction experiments, complemented by quantum chemical calculations to find and validate a structural model of the crystal form of the investigated molecule. This contribution will show the basics of NMR crystallography, its strong and weak points, a variety of its different flavours in which it is currently used, as well as its various applications.
Customised solutions in magnetic resonance: special probes for mas and in situ NMR on energy storage materials
Oliver Pecher1,2
1NMR Service GmbH, Blumenstr. 70 Haus 3, 99092 Erfurt, DE
2University of Cambridge, Yusuf Hamied Department of Chemistry, Lensfield Rd., Cambridge CB2 1EW, UK
The manifold applications of magnetic resonance include the research of new and innovative materials. NMR SERVICE develops and manufactures customised probes and accessories for solid-state NMR/NQR experiments and related methods. We guide you all the way from planning the setup, installing the hardware, supporting your experiments, troubleshooting, and hardware modifications/upgrades. Here, we will report on recent developments regarding MAS power handling and increased
experimental efficiency in solid-state NMR investigations, e.g.
- MAS 3.2 600 WB NMR probe optimised for low-gamma 39K NMR experiments
- Static in situ/operando NMR probes to research energy storage materials (Fig. 1)[1,2,3,4]
- Static in situ/operando NMR probe to measure coin cells
Figure 1. In situ NMR applied in battery research. Customised hardware and experimental approaches by NMR SERVICE help to make the experiments happen.
Furthermore, we will give some insights into our probes/accessories that combine the application of static solid-state NMR with electrochemical[1,2,3,4] and optical measurements (e.g. UV/Vis, IR/RA). Finally, the challenges and some solutions in 7Li and 23Na in situ NMR experiments on SiO[5] and Sn[6] anodes as well as hard carbon[7] are briefly discussed.
REFERENCES:
[1] K. Märker et al. J. Am. Chem. Soc. 2020, 142, 17447−17456. DOI: 10.1021/jacs.0c06727
[2] O. Pecher & D. M. Halat et al. J. Magn. Reson. 2017, 275, 127-136. DOI:10.1016/j.jmr.2016.12.008
[3] O. Pecher et al. Chem. Mater. 2017, 29, 213-242. DOI: 10.1021/acs.chemmater.6b03183
[4] O. Pecher et al. J. Magn. Reson. 2016, 265,200-209. DOI: 10.1016/j.jmr.2016.02.008
[5] K. Kitada et al. J. Am. Chem. Soc. 2019, 141(17), 7014-7027. DOI:10.1021/jacs.9b01589
[6] J. M. Stratford et al. J. Am. Chem. Soc. 2017, 139 (21), 7273-7286. DOI:10.1021/jacs.7b01398
[7] J. M. Stratford et al. Chem. Commun. 2016, 52, 12430-12433. DOI: 10.1039/C6CC06990H
Poster abstracts:
1. Electric Field Driven Surface Potential Tunability Probed by Scanning Probe Microscopy: An Effective Way to Tune the Surface Potential of the Polymers
Anand Babu
Reliance Jio Infocomm Ltd, Delhi, India
Surface Potential has prime importance in diverse fields of science, from cell adhesion to contact electrification. Conventionally, the techniques like surface chemical functionalization, ion injection, layer-by-layer deposition, and plasma etching are used to tune the surface potential of a material. However, these are complex multistep processes that need sophisticated techniques; moreover, long-time surface potential retentivity is also one of the challenging tasks here. We have proposed an effective single-step method to tune the surface potential of material by just reversing the direction of the electric field in an electrospinning setup, a nano/micro-fiber production unit where in situ electrical poling is happening during the production of the fibers, thus provides the high-stability in long-time retentivity of the surface potential. An atomic force microscopy-based non-contact scanning kelvin probe microscopy has been utilized to investigate the surface potential. Furthermore, we have observed that our proposed method is implementable to all the polymers, which are subject to producing the fibers in the electrospinning unit.
2. Cross-correlated spin relaxation in NMR studies of proteins
Paulina Bartosińska-Marzec1, Bartłomiej Banaś1, Clemens Kauffmann2, Robert Konrat2, Anna Zawadzka-Kazimierczuk1.
1Biological and Chemical Research Centre, Faculty of chemistry, University of Warsaw, ul. Żwirki i Wigury 101, 02-089 Warsaw, Poland
2University of Vienna, Vienna BioCenter, Campus-Vienna-Biocenter 5, 1030 Vienna, Austria
Intrinsically disordered proteins (IDPs) are highly dynamic proteins which play key roles in signaling and regulation of biological processes. Importantly, IDPs are not random coils, but transiently adopt certain structural motifs, related to their functions.[1] Thus their residual structure is worth studying. Nuclear magnetic resonance (NMR) is the only method which provides such information at atomic scale. Nonetheless, high dynamics leads to fast changes of the chemical environments of nuclei, which leads to averaging of chemical shifts, hampering the spectral analysis. The solution to this problem is using methods of high dimensionality, 4D and 5D, which reduces spectral crowding.[2]
One of the structural parameters that can be determined with NMR is dihedral angles distribution of the protein's backbone. For this we can use cross-correlated relaxation (CCR) rates. They are determined separately for individual amino acid residues and are calculated using the peak intensities.[3] Several types of CCR constants are required to determine the preferred conformation of individual residues.
We have developed seven 4D experiments to measure different CCR rates and successfully validated them on a well-known protein, ubiquitin (PDB code 1d3z).
Right now, we are conducting the measurements of an IDP, osteopontin.
Acknowledgments:
This work was supported by a grant from the Polish National Science Centre (MAESTRO, 2015/18/A/ST4/00270).
REFERENCES:
[1] P. Tompa, FEBS Lett., 2005, 15, 3346–3354
[2] K. Grudziąż, A. Zawadzka-Kazimierczuk, and W. Koźmiński, Methods, 2018, 148, 81–87
[3] C. Kauffmann, Zawadzka-Kazimierczuk A., Kontaxis R., Konrat R., ChemPhysChem. 2020, 21, 1
3. Improvement of cancer contrast in MRI using double action nanoparticles in the mouse animal model
Barbara Blasiak1,2, Armita Dash3, David MacDonald1, Frank C.J.M. van Veggel3, Boguslaw Tomanek1,2,4
1Polish Academy of Sciences, Institute of Nuclear Physics, Krakow, Poland
2Department of Clinical Neurosciences, University of Calgary, Calgary, Canada
3Department of Chemistry, University of Victoria, British Columbia, Canada
4Department of Oncology, University of Alberta, Edmonton, Canada
Magnetic Resonance Imaging (MRI) has been used for early cancer detection and treatment monitoring due to its excellent soft tissue contrast, yet in clinical settings contrast agents are used for cancer diagnosis. While MRI contrast may be provided solely by tissues themselves, due to differences in their relaxation times, contrast agents shortening T_1 and T_2 relaxation times further improve detection of small pathologies such as early stages breast cancers [1,2].
To improve the tumor contrast we have developed core/shell NaDyF4/NaGdF4 nanoparticles changing both T1 and T2 relaxation times of surrounding water molecules and conjugated them with tumor specific antibodies and proteins. The relaxation times (T1 and T2) of the nanoparticles with various score/shell sizes and concentrations were measured at 9.4T and 3T to find the optimum T1/T2 ratio for MRI. T1- and T2-weighted images using core/shell nanoparticles of the animal models of brain, breast and prostate cancer were collected and combined to provide enhanced contrast and edges. The contrast agents consisting of the core/shell nanoparticles with the optimal core and shell sizes are being developed to provide improved tumor contrast when the T1 and T2-weighted MR pulse sequences are applied.
Acknowledgments:
This work was funded by the National Science Center, Poland. Grant numbers: Harmonia no. 2018/30/M/NZ5/00844
REFERENCES:
[1] Blasiak B, Tomanek B, et al Detection of T2 changes in an early mouse brain tumor. Mag Res Imag 2010, 28:784-9.
[2] Tomanek B, Iqbal U, Blasiak B, at al. Evaluation of Brain Tumor Vessels Specific Contrast Agents for Glioblastoma Imaging. Neuro-Oncology, 2012, 14(1):53-63.
4. NMR spectroscopy – a versatile tool for GPx mimetic studies
Roxana-Alexandra Butuza, Anca Silvestru
Supramolecular Organic and Organometallic Chemistry Centre, Department of Chemistry, Faculty of Chemistry and Chemical Engineering, Babeş‐Bolyai University, 11 Arany Janos, RO‐400028, Cluj‐Napoca, România
In recent years an increased interest was given to organoselenium compounds containing organic groups with pendant arms capable for N→Se intramolecular interactions, due to their increased stability and improved properties such as enzymatic mimics, e.g. for Glutathione peroxidase (GPx).[1] As a continuation of our previous studies on diorganoselenium(II) compounds containing organic groups with pendant arms and/or pyrazole moieties,[2-4] we employed UV-Vis spectroscopy to investigate their antioxidant behavior via the Tomoda method.[5,6] In order to investigate the reaction mechanism of the compounds in the considered system (reaction between tiophenol and hydrogen peroxide), we employed multinuclear NMR spectroscopy. Given that the chemical shift in the 77Se NMR spectra has specific values for certain classes of compounds, 77Se NMR spectroscopy is an useful tool for the characterization of organoselenium compounds.[7] The obtained results show that the reactions occur via selenoxide intermediates which are stabilized by intramolecular N→Se interactions and such a behavior was sustained by DFT calculations as well.
Acknowledgments:
Financial support of UEFISCDI project no. PN-III-P4-ID-PCE-2020-1028 is highly acknowledged.
REFERENCES:
[1] A. J. Mukherjee, S. S. Zade, H. B. Singh, R. B. Sunoj, Chem. Rev. 110 (2010) 4357-4416.
[2] B. Danciu, R. A. Popa, A. Pop, V. Zaharia, C. Silvestru, A. Silvestru, Studia UBB Chemia LXI 3 (2016) 19-28.
[3] R. A. Popa, E. Licarete, M. Banciu, A. Silvestru, Appl. Organomet. Chem. 32 (2018) e4252.
[4] R. A. Popa, V. Lippolis, A. Silvestru, Inorg. Chim. Acta 520 (2021) 120272.
[5] M. Iwaoka, S. Tomoda, J. Am. Chem. Soc. 116 (1994) 2557-2561.
[6] V. Nascimento, N. L. Ferreira, R. F. S. Canto, K. L. Schott, E. P. Waczuk, L. Sancineto, C. Santi, J. B. T. Rocha, A. L. Braga, Eur. J. Med. Chem. 87 (2014) 131-139.
[7] H. Duddeck, Prog. Nucl. Magn. Reson. Spectrosc. 27 (1995) 1-323.
5. NMR sugar profile of honey from endangered bee species
Dessislava Gerginova, Svetlana Simova
Institute of Organic Chemistry with Centre of Phytochemistry – Bulgarian Academy of Sciences, Acad. G. Bonchev str., bl. 9, 1113 Sofia, Bulgaria
Bees are winged insects, known for their role in pollination. Over 20000 bee species are known in the world, only less than 10 are honey bees. Most common honey bee is Apis mellifera, which is mostly found in Europe, Africa, and Asia. Numerous bee species found in tropical and subtropical regions, primarily in South America, Africa, and Australia, are the stingless bees. While the honey bees belonging to the genus Apis comprise ~ 44 recognized subspecies, more than 550 species of stingless bees are e.g. included in tribe Meliponini. All share the ability to make honey and wax combs. A lot of information about specific behavior and characteristics is well known, however, the sugar composition of honeys produced by diverse genus and tribes is only poorly studied so far.
Honeys from four different Apis mellifera subspecies (A. m. ligustica – from Italy, A. m. mellifera – from Germany, A. m. ruttneri – from Malta, A. m. rodopica – from Bulgaria) and from two stingless bee species (Meliponula ferruginea – from Tanzania and Scaptotrigona Mexicana – from Mexico), were analysed using NMR spectroscopy. Their saccharide profile was determined by 13C spectra and compared using principle component and cluster analysis. Both methods show high potential to distinguish honeys from various bee species based on distinctive differences in the sugar compositions thus providing additional opportunities for consumer inclination to correct designation of natural food products.
Acknowledgments:
This research was funded by the Project BG05M2OP001-1.002-0012-C01: Centre of Competence „Sustainable utilization of bio-resources and waste from healing and aromatic plants for innovative bioactive products“, financed by Operational program „Science and education for smart growth“, co-financed by the European Union through the European structural and investment funds.
6. Investigating the miscibility of thermodynamically immiscible polymers by solid-state NMR
Iurii Vozniak, Ramin Hosseinnezhad, Sławomir Kaźmierski, Alina Vozniak, Andrzej Galeski
Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences, Lodz, Poland
The phase mixing efficiency of ABS and PET before and after SPD was investigated by 1H spin-lattice relaxation time measurements both in the laboratory and in rotating frames (T1, T1p) using solid-state 13C cross-polarization magic-angle spinning NMR experiments. NMR data were obtained using the Bruker AVANCE III WB 400 NMR spectrometer.
It is well known that each phase in immiscible polymer blend with phase separation has a unique characteristic 1H relaxation time. The relaxation time is almost the same for each polymer phase in miscible polymer blends that exhibit nanoscale phase mixing. According to the two polymer components, the original ABS/PET mixture displays two different 1H spin-lattice relaxation times in the laboratory frame (T1). On the other hand, the T1 values for the ABS and PET phases in the ABS/PET blend after severe plastic deformation processing are rather close. This could mean that the polymer mixing takes place within the interphase at the nanoscale, or in a range of tens of nanometers.
Acknowledgments:
This research was funded in part by National Science Centre (Poland) under the grant 2021/43/B/ST8/01443.
7. Simultaneous Reversible and Irreversible Parahydrogen NMR Signal Ehancement
Sylwia Jopa1, Adam Mames2, Mariusz Pietrzak2 and Tomasz Ratajczyk2
1Faculty of Chemistry, University of Warsaw, Pasteura 1, Warsaw 02-093, Poland
2Institute of Physical Chemistry, Polish Academy of Sciences, Kasprzaka 44/52, Warsaw 01-224, Poland
Although Magnetic resonance (MR) is widely employed as a powerful non-invasive research tool, it suffers from poor sensitivity, limiting further MR applications.[1,2] Hyperpolarization (HP) techniques can significantly enhance the MR signal.[3] The most prospective HP methods are irreversible hydrogenative Parahydrogen Induced Polarization (hPHIP) and Signal Amplification by Reversible Exchange (SABRE).[4] These two methods are complementary to each other, as SABRE can hyperpolarize some molecules which hPHIP cannot, and vice versa. The methods are based on the same source of hyperpolarization -parahydrogen. In this context, the general question is how to combine SABRE and hPHIP, what will happen when they are combined and how the interplay between them can be manipulated. Therefore, we have investigated the issue of the combination of hPHIP and SABRE.[5]
As a molecule for SABRE-hPHIP 3-ethynylpyridine was chosen because it contains two fragments: pyridine and acetylene units. Pyridine unit can undergo SABRE, while the triple bond can by hydrogenated -thus, hPHIP is possible. As a catalyst, [Ir(COD)(IMes)Cl] (where IMes is the N-heterocyclic carbene ligand = 1,3-bis(2,4,6-trimethyl-phenyl)imidazol-2-ylidene and COD = 1,5-cyclooctadiene), was used as this catalyst is used for SABR[6] and can also facilitate hydrogenation[7].
In our experiments, we observed simultaneous SABRE and hPIPH. As time progressed, both effects were step-by-step suppressed. The possible factors which caused the suppression of hPHIP and SABRE were discussed. In particular, the possible scenarios of the deactivation of the catalysts were presented. We have also observed that the reaction mechanism of simultaneous SABRE and hPHIP hyperpolarization could be modulated by the initial activation of the catalyst with a co-ligand. In particular, the hPHIP effect can be suppressed entirely, but stable and efficient SABRE in return was observed.
To conclude, hPHIP-SABRE is possible, and the interplay between hPHIP and SABRE can be modulated. However, further research is required to understand hPHIP-SABRE more comprehensively, which can facilitate the utilization of hPHIP-SABRE in medical diagnostics and catalysis.
Acknowledgments:
This work was financially supported by the National Science Centre in Poland (OPUS 11 2016/21/B/ST4/02162, SONATA-2011/03/D/ST4/02345).
REFERENCES:
[1] J. H. Lee, Y. Okuno, S. Cavagnero, J. Magn. Reson. 2014, 241, 18–31.
[2] J.-H. Ardenkjaer-Larsen, G. S. Boebinger, A. Comment, S. Duckett, A. S. Edison, F. Engelke, C. Griesinger, R. G. Griffin, C. Hilty, H. Maeda, G. Parigi, T. Prisner, E. Ravera, J. van Bentum, S.Vega, A. Webb, C. Luchinat, H. Schwalbe, L. Frydman, Angew. Chem., Int. Ed. 2015, 54, 9162.
[3] K. Münnemann, H. W. Spiess, Nat. Phys. 2011, 7, 522–523.
[4] K. V Kovtunov, E. V Pokochueva, O. G. Salnikov, S. F. Cousin, D. Kurzbach, B. Vuichoud, S. Jannin, E. Y. Chekmenev, B. M. Goodson, D. A. Barskiy, I. V Koptyug, Chem. – An Asian J. 2018, 13, 1857–1871.
[5] A. Mames, S. Jopa, M. Pietrzak, T. Ratajczyk, RSC Adv. 2022, 12, 15986–15991.
[6] M. J. Cowley, R. W. Adams, K. D. Atkinson, M. C. R. Cockett, S. B. Duckett, G. G. R. Green, J. A. B. Lohman, R. Kerssebaum, D. Kilgour, R. E. Mewis, J. Am. Chem. Soc. 2011, 133, 6134.
[7] V. Nesterov, D. Reiter, P. Bag, P. Frisch, R. Holzner, A. Porzelt, S. Inoue, Chem. Rev. 2018, 118, 9678–9842.
8. Solution behavior and solid state structure of new di- and triorganolead(IV) complexes with tetraphenyldichalcogenoimidodiphosphinato ligands
Eleonóra Kapronczai, Cristian Silvestru
Supramolecular Organic and Organometallic Chemistry Centre, Faculty of Chemistry and Chemical Engineering, Babeș-Bolyai University,11 Arany János Street, 400028 Cluj-Napoca, Romania
Tetraorganodichalcogenoimidodiphosphinato ligands exhibit a large variety of coordination patterns,[1] but only a few studies on the synthesis and structural characterization of organolead(IV) complexes with such ligands were reported so far.[2-4] Single-crystal X-ray diffraction studies on dimethyl- and diphenyllead(IV) derivatives with imidodiphosphinato ligands revealed an O,O-bidentate coordination to lead,[3,4] thus leading to six-membered inorganic chelate rings. In order to investigate the structure of organolead(IV) complexes and the coordination behavior of this type of ligands, new di- and triorganolead(IV) compounds with tetraphenyldichalcogenoimidodiphosphinato ligands were synthetized. The new compounds were characterized in solution by multinuclear NMR spectroscopy and for several of them the solid state structure was determined by single crystal X-ray diffraction. For selected compounds the dynamic behavior in solution was investigated by variable temperature NMR experiments. The decomposition process of the triorganolead(IV) derivatives was studied by time-related NMR experiments.
Acknowledgments:
This work was supported by the Romanian Ministry of Education and Research (PN-III-P4-ID-PCCF-2016-0088).
REFERENCES:
[1] C. Silvestru, J. E. Drake, Coord. Chem. Rev. 223 (2001) 117-216.
[2] R. A. Varga, C. Silvestru, I. Haiduc, Synth. React. Inorg. Met.-Org. Chem. 30 (2000) 485-498.
[3] R. Varga, J. E. Drake, C. Silvestru, Phosphorus, Sulfur Silicon Relat. Elem. 169 (2001) 47-50.
[4] R. A. Varga, J. E. Drake, C. Silvestru, J. Organomet. Chem. 675 (2003) 48-56.
9. Linezolid co-crystals structure elucidation by joint use of high-resolution solid state NMR and crystal structure prediction (CSP) calculations
Mehrnaz Khalaji, Piotr Paluch, Marta K. Dudek, Marek J. Potrzebowski
Center of Molecular & Macromolecular Studies, Polish Academy of Sciences, Sienkiewicza 112, 90-363 Lodz, Poland
Mechanococrystalization of linezolid, a wide-range antibiotic, results micro-crystalline powders, in some of them makes impossible the application of single crystal X-Ray diffraction in structural elucidation.[1,2] We expect a joint use of high-resolution solid-state NMR and crystal structure prediction (CSP) calculations can be successful. However, linezolid has conformational freedom by Having four rotatable bonds, some of which can assume many discreet values poses a challenge in establishing its conformation in a solid phase also the CSP-NMR protocol meets serious obstacles, including ambiguities in NMR signal assignment.[2,3] We demonstrate a possible way of avoiding these obstacles and making as much use of conformations of LIN inside the crystal. CSP calculations yield model crystal structures examined against experimental solid-state NMR data, leading to a reliable identification of the most probable molecular arrangement. Our work led to the crystal structure elucidation of a co-crystal of linezolid (LIN) with 2,3-dihydroxybenzoic acid, however, the challenging case of a co-crystal of LIN with 2,4-dihydroxybenzoic acid led to the identification of the most probable conformations of LIN inside the crystal.[3]
Acknowledgments:
This work was financially supported by Polish National Science Centre under Sonata 14 grant No. UMO-2018/31/D/ST4/01995. The Polish Infrastructure for Supporting Computational Science in the European Research Space (PL-GRID) is gratefully acknowledged for providing computational resources. A purchase of a Panalytical powder X-Ray diffractometer used to obtain results included in this publication was supported by funds from the EU Regional Operational Program of the Lodz Region, RPLD.01.01.00-10-0008/18.
REFERENCES:
[1] M. Khalaji, M. K. Dudek, M. J. Potrzebowski, Cryst. Growth. (2021) 2301–2314.
[2] M. Khalaji, A. Wróblewska, E. Wielgus, G. D. Bujacz, M. K. Dudek, M. J. Potrzebowski, Acta Cryst. 76 (2020) 892−912.
[3] M. Khalaji, P. Paluch, M. K. Dudek, M. J. Potrzebowski, Solid State Nuclear Magnetic Resonance. 121 (2022) 101813.
10. dGAE (297-391) tau filaments seen by solid-state NMR
Kristine Kitoka
Tauopathies are the most troublesome of all age-related chronic conditions, as the development of therapies has an extremely low success rate compared to other chronic diseases. Tau protein is a common molecular denominator of tauopathies, which is found as insoluble deposits in specific brain parts. Pathological tau is a promising molecular target to tackle. However, the field still lacks a deeper understanding of the structural details of tau filaments. Recently, it has been shown that truncated tau construct dGAE (297-391) can form filaments that closely resemble paired helical filaments derived from Alzheimer's disease brain tissue (Al-Hilaly et al., 2017; Al-Hilaly et al., 2019; Lovestam et al., 2022).
Here, we aim to determine the global fold dGAE tau filaments using solid-state NMR. Moreover, we aim to investigate whether chemical shifts of dGAE and seeded by dGAE full-length tau filaments differ.
Double-labeled proteins were expressed in E. coli and purified using a combination of CEX, IMAC, and SEC. A set of 2D and 3D solid-state experiments was recorded to assign secondary structures and investigate the global fold of filaments. The macroscopic features of the tau filaments were also examined by AFM and negative stain EM and complimented with various biophysical experiments.
11. The assessment of the distribution of drug-loaded polyelectrolyte nanocarriers by the 1H or 19F Magnetic Resonance Imaging
Natalia Łopuszyńska1, Krzysztof Szczepanowicz2, Kamil Stachurski1, Marta Szczęch2, Krzysztof Jasiński1, Władysław P. Węglarz1
1Institute of Nuclear Physics Polish Academy of Sciences, Cracow, Poland
2Institute of Catalysis and Surface Chemistry Polish Academy of Sciences, Cracow, Poland
The main disadvantages of chemotherapy (or more generally pharmaceutical therapies) include toxic side effects and the development of resistance to the chemical agents, which is related to the non-specific targeting of therapeutic agents. Although novel approaches are constantly proposed, there are still many obstacles to overcome. Among drug delivery systems, polymeric nanocarriers for passive targeting based on the EPR effect are intensively investigated. Modification of such nanocarriers with imaging agents introduces the possibility of the observation of drugs kinetics and the assessment of the efficiency of the therapeutics transport to the site of action. Among various available imaging modalities, Magnetic Resonance Imaging (MRI) has become a powerful tool for non-invasive imaging due to its high spatial resolution and its inherent safety. The present contribution aimed to investigate the possibility of detection of new theranostic polyelectrolyte nanocarriers by standard, proton magnetic resonance imaging, as well as 19F MRI.
Various types of nanocarriers were formed via the layer-by-layer technique, i.e. by the deposition of ionic and cationic polyelectrolyte shell layers on nanoemulsion drops. MRI contrast agents were either embedded in their shells or encapsulated in cores. In particular, small particles of iron oxide (Fe3O4) as were used negative type; Gadolinium ions as positive type, and NafionTM polymer or fluorouracil (FU) as hot-spot type of MRI contrast agents. Both the 1H and 19F MR spectroscopy/relaxation measurements, as well as 1H and 19F imaging, were performed at the 9.4 T BioSpec 94/20 preclinical MRI scanner (Bruker BioSpin, Germany), equipped with BGA60-S gradient coils and 35 mm birdcage RF coil, controlled by Paravision 5.1 software. Additionally, a small transmit-receive ribbon solenoid RF coil (ID of 14 mm), which can be tuned either to 1H or 19F resonant frequency was used for 19F experiments.
The presented results prove the distribution of drug-loaded polyelectrolyte nanocarriers can be reliably assessed by the 1H or 19F MRI. Due to excellent contrasting properties, proved by high values of molar relaxivities r1 and r2, polyelectrolyte nanocapsules with contrast agents of relaxation type (Fe3O4 and Gd) should by suitable for the use in standard clinical MRI setting. However, due to close to zero natural background, the use of 19F MRI as the detection method presents an interesting alternative to classic 1H-MRI with relaxation-based contrast agents by providing excellent contrast and enabling straightforward quantification of the drug delivery. Regardless of the unfavourable characteristic of NafionTM for the MR imaging, i.e. multiple resonances in MR spectrum and very short T2 time, it was possible to reliably visualize nanocarriers’ distribution in the sample. As the fluorouracil has an NMR spectrum with only one resonance line and relatively long T2 relaxation, better SNR was achieved within the same acquisition time. This allowed us to shorten the acquisition time to as low as 4 minutes while keeping SNR high enough. From the perspective of the potential preclinical and clinical applications, this suggests the possibility of in-vivo tracking at a 5-FU therapeutic dose.
Acknowledgments:
This work was partially financed from the Norwegian Financial Mechanism, GRIEG Project 2019/34/H/ST5/00578
12. Harvest time affects antioxidant capacity, total polyphenol and flavonoid content of Polish St John’s wort’s (Hypericum perforatum L.) flowers
Katerina Makarova1, Joanna J. Sajkowska‑Kozielewicz1, Katarzyna Zawada1, Ewa Olchowik‑Grabarek2, MichałAleksander Ciach3,4, KrzysztofGogolewski3, Natalia Dobros1, Paulina Ciechowicz1, Hélène Freichels5, Anna Gambin3
1Department of Physical Chemistry, Chair of Physical Pharmacy and Bioanalysis, Faculty of Pharmacy With Laboratory Medicine Division, Medical University of Warsaw, Banacha 1, 02‑097 Warsaw, Poland.
2Department of Microbiology and Biotechnology, Faculty of Biology, University of Bialystok, Ciolkowskiego 1J, 15‑245 Bialystok, Poland.
3Faculty of Mathematics, Informatics and Mechanics, University of Warsaw, Stefana Banacha 2, 02‑097 Warszawa, Poland.
4Centre for Statistics, Hasselt University, Diepenbeek, 3590 Limburg, Belgium.
5Magritek GmbH, Philipsstraße 8, 52068Aachen, Germany.
The polyphenol content and antioxidant capacity of hyperforin and hypericin-standardized H. perforatum L. extracts may vary due to the harvest time. In this work, ethanol and ethanol–water extracts of air-dried and lyophilized flowers of H. perforatum L., collected throughout a vegetation season in central Poland, were studied. Air-dried flowers extracts had higher polyphenol (371 mg GAE/g) and flavonoid (160 mg CAE/g) content, DPPH radical scavenging (1672 mg DPPH/g), ORAC (5214 µmol TE/g) and FRAP (2.54 mmol Fe2+/g) than lyophilized flowers extracts (238 mg GAE/g, 107 mg CAE/g, 1287 mg DPPH/g, 3313 µmol TE/g and 0.31 mmol Fe2+/g, respectively). Principal component analysis showed that the collection date influenced the flavonoid and polyphenol contents and FRAP of ethanol extracts, and DPPH and ORAC values of ethanol–water extracts. The ethanol extracts with the highest polyphenol and flavonoid content protected human erythrocytes against bisphenol A-induced damage. Both high field and benchtop NMR spectra of selected extracts revealed differences in composition caused by extraction solvent and raw material collection date. Moreover, we have shown that benchtop NMR can be used to detect the compositional variation of extracts if the assignment of signals is done previously
13. Solid state NMR investigation of CO2 adsorption on NaX zeolite
Momchil Momchilov1, Yavor Mitrev1 , Pavletta Shestakova1*, Peter Georgiev2*
1Institute of Organic Chemistry with Centre of Phytochemistry, Bulgarian Academy of Sciences, Acad. G. Bonchev Str., Bl. 9, 1113
2Condensed Matter Physics and Microelectronics Department, Faculty of Physics, University of Sofia, J. Bourchier 5, Sofia 1164, Bulgaria
The momentum-gaining drive towards sustainable green society requires a dramatic reduction in the emission of industrial and household gases, such as CO2 . Various methods and materials have been considered and are under development, for CO2 capture and storage. Faujasite type zeolites (FAU) have been considered as one of the most promising microporous materials for CO2 capture, due to their relatively large capacity, durability and mainly – high CO2 binding energies in excess of 40-50 kJ/mol. The heat of adsorption become reduced by factors of, e.g. 2- 3, as the captured CO2 amount in the zeolite is increased, thus limiting the overall useful capacity of the material under realistic operational conditions. It is therefore of high practical and fundamental interest to elucidate the particular coordination environments responsible for the high binding energy. In the present study we used solid state NMR to investigate the microscopic aspects of 13CO2 adsorption in pure FAU microcrystalline phase NaX zeolite, prepared from refined natural kaolin by own-synthesis procedure. Samples containing different 13CO2 amounts were prepared exposing the zeolite to different equilibrium 13CO2 pressures, spanning wide range of the material adsorption capacity, at room temperatures. Two types of NMR experiments were performed to determine the nature of the adsorbed CO2 - chemisorbed or physically adsorbed. The 1H-13C CPMAS NMR experiments lead to a selective increase of the signal of chemisorbed 13CO2 due to possibility for transfer of magnetization from neighboring protons of the zeolite matrix as well as due to its immobilization upon its coordination to Na+ ions. These experiments are not suitable for registration of physically adsorbed 13CO2, since the CP transfer is inefficient due to its higher mobility. To detect the presence of physisorbed 13CO2 , 13C spectra with high power proton decoupling were measured.
The results show that at low CO2 pressure (24 mbar) only one type of chemisorbed CO2 species are present and the quantitative ratio of chemi- to physisorbed CO2 species is approximately 1:1. At higher pressures the complexity of the chemisorbed CO2 resonance increases indicating additional binding of the CO2 molecules to Na + ions located at different positions in the zeolite matrix. The 23Na spectra demonstrate complex spectral pattern due to the presence of at least 4 different nonequivalent Na sites in the zeolite. Comparison of the line shapes of 23Na spectra of pure NaX and samples with different CO2 pressure further suggests the possibility for different binding modes between CO2 and Na+ ions upon increase of the CO2 loading. The 29Si and 27Al NMR spectra of the studied samples show only minor differences indicating that the zeolite structure is not influenced by the CO2 loading.
Acknowledgments:
This work was supported by the Bulgarian Ministry of Education and Science under the National Research Programme E+: Low Carbon Energy for the Transport and Households, grant agreement D01-214/2018 and INFRAMAT Project (Research equipment of Distributed Research Infrastructure), part of Bulgarian National Roadmap for Research Infrastructures.
14. 13C CP MAS NMR spectroscopy in fake drug analysis
J. Harwacki, Ł. Szeleszczuk, D.M. Pisklak
Department of Physical Chemistry, Faculty of Pharmacy, Warsaw Medical University, Warsaw, Poland
Counterfeit medicines are a growing worldwide problem. According to Global Surveillance and Monitoring System and the World Health Organization (WHO)it is estimated that about 10% of all medical products used in low- and middle-income countries are substandard and falsified(WHO, 2017), but in the last decade this problem has been growing and now affects also highly developed countries. There are several groups of pharmaceutical products that are most commonly falsified. .In the case of highly developed countries, preparations for erectile dysfunction are among the most frequently counterfeited medicinal products which, thanks to illegal distribution, could be easily available without the need for medical consultation. From a pharmaceutical point of view, counterfeit drugs pose a health risk due to the possible content of API different from the declared one, differences in the kinetics of drug release, as well as the possible presence of toxic substances. Therefore, development of innovative analytical approaches for falsified drug identification is essential.
The main aim of the study was to assess the possibility of using 13C CP MAS NMR spectroscopy to identify falsified drugs.In our study we showed that 13C CPMAS NMR spectroscopy allows for easy and unambiguous identification of counterfeit products. However, the practical applicability of this technique is limited due to the limited availability of equipment and the cost of measurement. Therefore, in a further stage of the work, the 13C CP MAS NMR spectrum pattern analysis allowed for the identification of structural differences which enabled the development of simple dyeing techniques making the identification of illegal products possible.
15. Linear discriminant analysis reveals hidden patterns in NMR chemical shifts of intrinsically disordered proteins
Paulina Putko
Centre of New Technologies, University of Warsaw
Nuclear magnetic resonance spectroscopy (NMR) is the key method to study intrinsically disordered proteins (IDPs). The standard assignment of signals in protein NMR spectra is based on analyzing a set of results from three-dimensional experiments, which provide information on the sequential linkage (correlation) of chemical shifts of individual nuclei, e.g., 3D HNCACB. Yet, even the first step of such analysis, i.e., assignment of observed resonances to particular nuclei, is often very difficult due to low peak dispersion in the spectra of IDPs.
We propose using the advanced statistical method in the assignment process, which can be effectively supported by finding 'hidden' chemical shift patterns, specific for the amino acid residue types. The patterns are found in the training data from Biological Magnetic Resonance Bank (BMRB) using linear discriminant analysis (LDA) and then used to classify spin systems in a fresh alfa-synuclein sample. We show that a procedure can greatly support the analysis of NMR spectra.
16. Solid state NMR characterization of novel polymer/calcium phosphate hybrid materials for biomimetic remineralization of tooth enamel
K. Ruseva1, M. Simeonov1, E. Vassileva1, E. Dyulgerova2, R. Ilieva3, R. Gergulova3, D. Rabadjieva3, P. Shestakova4
1Laboratory on Structure and Properties of Polymers, Faculty of Chemistry and Pharmacy, University of Sofia, 1, James Bourchier blvd., 1164 Sofia, Bulgaria,
2Faculty of Dental Medicine, Medical University, 1, G. Sofiiski Str., 1431 Sofia, Bulgaria,
3Institute of General and Inorganic Chemistry, Bulgarian Academy of Sciences, 1113 Sofia, Bulgaria,
4Bulgarian NMR Centre, Institute of Organic Chemistry with Centre of Phytochemistry, Bulgarian Academy of Sciences, Acad. G. Bonchev Str., Bl. 9, 1113 Sofia, Bulgaria,
Development of new materials for biomimetic remineralization of tooth enamel is a topic of great importance in the dental materials research as a new approach to the prevention, restoration and treatment of defective enamel. Recent studies demonstrated that polymers of appropriate functionality can play a role in controlling mineralization reactions and lead to the desired calcium phosphate phase or mixture of phases.
In the present study novel Polymer/Calcium Phosphate (P/CaP) hybrid materials were synthesized and characterized by solid state NMR spectroscopy. The hybrid P/CaP systems were obtained by in situ precipitation of CaP in a solution of the polymers under different reaction conditions. For comparison pure CaP synthesized under identical conditions as for the preparation of hybrid systems were also investigated. 1H, single pulse 31P and 1H®31P cross-polarization (CP) MAS solid state NMR spectra provided insight to the structural characteristics and chemical composition of calcium phosphate phase formed in studied materials, as a function of sample preparation procedure and synthesis conditions. The NMR data revealed that the pure CaP phase formed under controlled conditions (pH, temperature, maturation time) represent a mixture of crystalline and poorly crystalline (nanocrystalline) hydroxyapatite (HAP). The 31P NMR spectrum of CaP phase formed in the absence of polymer under identical conditions as for the synthesis of the hybrid materials, however, represented a complex pattern indicating the presence of mixture of orthophosphates and hydrogen phosphate species. The NMR spectra of the hybrid polymer/CaP sample obtained by in situ precipitation in a solution of polymer demonstrate the formation of amorphous calcium phosphate, clearly evidencing the role of the polymer on the composition and structural properties of the CaP phase.
Acknowledgments:
The financial support of the National Science Fund under project КP-06-Н49-6/2020 is gratefully acknowledged. Research equipment of Distributed Research Infrastructure INFRAMAT, part of Bulgarian National Roadmap for Research Infrastructures, supported by Bulgarian Ministry of Education and Science was used in this investigation.
17. Analysis of the composition of biodiesel obtained from the esterification of rapeseed oil and goose fat
Krzysztof Siczek1, Marek Woźniak1, Mateusz Zakrzewski2, Tomasz Bartosik3
1Department of Vehicles and Fundamentals of Machine Design, Lodz University of Technology, Poland
2Institute of General and Ecological Chemistry, Lodz University of Technology, Poland
3Institute of Organic Chemistry, Lodz University of Technology, Poland
To reduce emissions, various biofuels are used as an additive to diesel fuel supplied to internal combustion engines, both automotive and stationary. The energetic properties of biofuels are strongly dependent on their chemical composition identified using various methods, including NMR spectroscopy, chromatography, and others. The present work utilized the 1H NMR and the 13C NMR spectroscopy using the Bruker Avance apparatus and the gas chromatography using a Hewlett Packard series 5890 chromatograph (Agilent Technologies) coupled with a Hewlett Packard series 5972 mass spectrometer. Two biodiesel samples were tested: the first one in the form of popular methyl ester of rapeseed oil and the other in the form of methyl ester of goose fat obtained via transesterification process for the first time in the world at the Lodz University of Technology. The obtained results showed that the methyl ester of goose fat showed the presence of a higher amount of saturated hydrocarbons than in the case of the methyl ester of rapeseed oil. Although the NMR spectrography turned out to be less accurate than gas chromatography, it also allowed specifying the basic components contained in both tested biodiesel samples.
18. 1H NMR in verification of hemp-seed oil quality
Paweł Siudem, Katarzyna Paradowska
Department of Physical Chemistry, Faculty of Pharmacy, Warsaw Medical University, Warsaw, Poland
Currently, there is a growing interest in the health-promoting effects of vegetable oils, used both in the food and pharmaceutical industries. Consumers pay attention to the high quality of the consumed oil, hence there is a constant search for methods enabling quick and routine quality control of vegetable oils. Hemp-seed oil, the main food product obtained from hemp, is most frequently produced by cold pressing of seeds. Hemp-seed oil exhibits health benefits due to its high content of essential fatty acids. It is composed in approximately 80% of polyunsaturated fatty acids (PUFA), mainly of ω-6 linoleic acid (LA) and ω-3 alpha-linolenic acid (ALA). The LA:ALA ratio is ca. 3:1, as it is recommended in healthy diet.
Six samples of cold pressed hemp-seed oils were studied by nuclear magnetic resonance (NMR) and Fourier-transformed infrared spectroscopy (FT-IR). These methods were applied to evaluate the ratio of ω-6 to ω-3 fatty acids and to control the quality of hemp oils. FT-IR method allowed us to distinguish three oils with the highest level of γ-linolenic acid. Additionally, NMR spectroscopy was applied to determine the ω-6 to ω-3 ratio. The outcomes of the NMR experiment are in agreement with GC outcomes measures. The results indicate that NMR and FT-IR may be used in routine evaluation of hemp-seed oil quality as a fast and reliable method of verification of ω-6 to ω-3 ratio and origin of the oil.
19. Structure and Conformational Mobility of 1,3,5-Triazine Derivatives for OLED Applications
Georgi Dobrikov1, Yana Nikolova1, Ivailo Slavchev1, Miroslav Dangalov1, Vera Deneva1, Luidmil Antonov2, Nikolay G. Vassilev1
1Institute of Organic Chemistry with Centre of Phytochemistry, Bulgarian Academy of Sciences, Acad. G. Bonchev Str., Bl. 9, 1113 Sofia, Bulgaria,
2Institute of Electronics, Bulgarian Academy of Sciences, 72, Tsarigradsko chaussee blvd., Sofia 1784, Bulgaria
Series of compounds consisting of 1,3,5-triazine core linked to various aromatic arms by amino group were synthesized and characterized. In solution the studied compounds exist as a mixture of two conformers: symmetric propeller and asymmetric conformer, in which the one aromatic arm is rotated around C-N bond. At temperatures below -60oC the VT NMR spectra in DMF-d7 are in slow exchange regime and the signals of two conformers were elucidated. At temperatures above 100oC the VT NMR spectra in DMSO-d6 are in fast exchange regime and the averaged spectra were measured. The populations of propeller and asymmetric conformers in DMF-d7 vary from 14:86 to 50:50 depending from the substituents. The rotational barriers of propeller and asymmetric conformers in DMF-d7 were measured for all compounds and are in the interval from 11.7 to 14.7 kcal/mol. The DFT calculations of structures of studied compounds either in ground states (conformers) and in transition states (the rotation around the C-N bonds) predict energy differences, which are in good agreement with the experimental rotational barriers. The DFT calculations reveal that the observed chemical exchange occurs by the rotation around the C(1,3,5-triazine)-N bond. The absorption and emission spectra of studied compounds were measured in acetonitrile, dichloromethane and methanol.
Acknowledgments:
The financial support by the Bulgarian Science Fund (UNA-17/2005, DRNF-02/13, КП-06-Н29/6) is gratefully acknowledged. Research equipment of Distributed Research Infrastructure INFRAMAT, part of Bulgarian National Roadmap for Research Infrastructures, supported by Bulgarian Ministry of Education and Science was used in this investigation.
20. DNA construct optimization for the NMR structural studies of the 8-17 DNAzyme
Julia Wieruszewska
DNAzymes are synthetic enzymes based on DNA, able to catalyze several chemical reactions – especially when substrates are nucleic acids. Thanks to their resistance to degradation, DNAzymes show promise for future use in biotechnology and medicine, as biosensors or therapeutics, e.g. for gene silencing. Unfortunately, their mechanisms of catalytic action has not yet been fully understood, hindering their practical use. The 8-17 DNAzyme, which is the main focus of our research, is able to catalyze the cleavage of RNA strands at precisely defined points in the sequence. It is a DNA metalloenzyme active in the presence of divalent metal ion cofactors such as Pb2+, Zn2+ or Mg2+. Surprisingly, the research into 8-17 structure using low resolution techniques, such as CD spectroscopy and FRET, revealed that it appears to possess two very distinct active conformations depending on the metal cofactor present1. Namely, while Pb2+ appears to bind directly to the apo-enzyme, interaction with other activating ions is accompanied by a large scale structural change, towards a more compact fold. When the crystal structures of the apo- and Pb2+-bound forms of 8-17 DNAzyme were finally obtained, they turned out to indeed be identical, while displaying an unexpectedly intricate network of tertiary interactions2. However, the second catalytically active conformation of the DNAzyme – induced by cofactors such as Zn2+ or Mg2+– remains elusive to crystallization.
In our research we set out to study the folding of 8-17 DNAzyme in the presence of Zn2+ using biomolecular NMR methods in solution. Given the large size of 8-17 DNAzyme constructs usually employed and molecular weight limitations of NMR, a careful optimization of the 8-17 DNAzyme construct to be studied turned out to be a necessary prerequisite for any such investigations. Through a process of gradual truncation and sequence-optimization of the DNAzyme’s target-binding arms we were able to arrive at a variant composed only 35 nucleotides (with the classic 8-17 constructs counting >65 nt), without affecting its enzymatic capabilities or metal binding, as assessed by gel electrophoresis and CD spectroscopy. The shortened construct turned out adopt a single NMR spectral form in the presence of Zn2+ with sharp lines allowing for resonance assignments and distance restraint derivation without isotope enrichment. Restrained MD simulations are now ongoing of elucidate the elusive Zn2+-bound form of the 8-17 DNAzyme.
Acknowledgments:
This work has been supported by the National Science Center (Poland) under grant agreement: [UMO-2018/31/D/ST4/01467] to W.A.. The calculations were performed at Poznań Supercomputing and Networking Center.
REFERENCES:
[1] K. Schlosser, and Y. Li “A Versatile Endoribonuclease Mimic Made of DNA: Characteristics and Applications of the 8–17 RNA-Cleaving DNAzyme”, ChemBioChem, 2010, 11, 866–879.
[2] H. Liu, X. Yu, Y. Chen, J. Zhang, B. Wu, L. Zheng, P. Haruehanroengra, R. Wang, S. Li, J. Lin, J. Li, J. Sheng, Z. Huang, J. Ma & J. Gan, “Crystal structure of an RNA-cleaving DNAzyme”, Nat. Commun., 2017, 8:2006
21. Topological preferences of a systematic set of model DNA G-quadruplexes studied by NMR and low resolution techniques
Amadeusz Woś
The principles underlying the folding of G-quadruplexes remain unexplained thus hindering the analysis of biological processes involving them, as well as, efforts toward their application in nanotechnology. The topological architecture of G4 structures depends on many factors and has not been fully understood [1]. The aim of our research is to understand the relationship between the G4 structures and their nucleotide sequences in order to allow for the design of G-quadruplexes with desired conformations. In order to study the relationship between loop length and conformational preferences we have systematically investigated a set of all 64 two-tetrads G4 containing thymidine–only loops of 1-4 length (GGT1-4GGT1-4GGT1-4GG). We applied a combination of biophysical tools (NMR, CD and UV-VIS spectroscopies) to characterize the G4 folds and native gel electrophoresis to determine the number of structures formed in each case. All 64 sequences form G-quadruplexes both in Na+ and K+ buffers, although their stabilities vary significantly. For sequences adopting a single form structural studies are ongoing.
22. The usage of chemometric methods and NMR spectroscopy in examination of dietary supplements containing Hawthorn fruit (Crataegus)
Hanna Dąbrowska1, Katarzyna Zawada1, Katerina Makarova1, Dariusz Gołowicz2, Krzysztof Kazimierczuk2
1Medical University of Warsaw, Faculty of Pharmacy, Department of Physical Chemistry
2University of Warsaw, Centre of New Technologies
The work aimed to investigate the efficacy of a combination of low-field NMR spectroscopy with chemometric analysis to identify and differentiate hawthorn species occurring in preparations containing hawthorn fruit or its extract, available on the Polish market.
1H NMR spectra of extracts from 14 preparations available on the Polish market containing hawthorn fruit were recorded using a benchtop NMR spectrometer (resonance frequency: 43 MHz). Despite the dominance in the spectra of signals from the solvent the spectra of different preparations showed visible variation. However, classical quantitative analysis with direct use of the spectra could not be performed in all cases due to the lack of visible signals in the phenolic range. PCA analysis established that it was possible to differentiate samples using the phenolic range (6-8 ppm) even in cases where the intensity of signals from the active components of hawthorn was low, while it was not possible to use other spectral ranges to reliably differentiate samples. However, also differentiation using only the polyphenol range did not allow for the classification of samples according to the species of hawthorn used in a given preparation.