New Article in Journal of Biomolecular NMR
Jerzy Sitkowski, Wojciech Bocian, Elżbieta Bednarek, Mateusz Urbańczyk, Wiktor Koźmiński, Piotr Borowicz, Grażyna Płucienniczak, Natalia Łukasiewicz, Iwona Sokołowska, Lech Kozerski
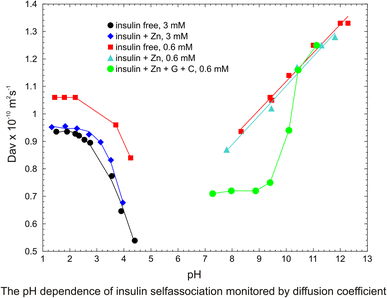
The NMR derived translational diffusion coefficients were performed on unlabeled and uniformly labeled 13C,15N human insulin in water, both in neat, with zinc ions only, and in pharmaceutical formulation, containing only m-cresol as phenolic ligand, glycerol and zinc ions. The results show the dominant role of the pH parameter and the concentration on aggregation. The diffusion coefficient Dav was used for monitoring the overall average state of oligomeric ensemble in solution. The analysis of the experimental data of diffusion measurements, using the direct exponential curve resolution algorithm (DECRA) allows suggesting the two main components of the oligomeric ensemble. The 3D HSQC-iDOSY, (diffusion ordered HSQC) experiments performed on 13C, 15N-fully labeled insulin at the two pH values, 4 and 7.5, allow for the first time a more detailed experimental observation of individual components in the ensemble. The discussion involves earlier static and dynamic laser light scattering experiments and recent NMR derived translational diffusion results. The results bring new informations concerning the preparation of pharmaceutical formulation and in particular a role of Zn2+ ions. They also will enable better understanding and unifying the results of studies on insulin misfolding effects performed in solution by diverse physicochemical methods at different pH and concentration.
|
New Article in Methods
Katarzyna Grudziąż, Anna Zawadzka-Kazimierczuk, Wiktor Koźmiński

Intrinsically disordered proteins (IDPs) are getting more and more interest of the scientific community. Nuclear magnetic resonance (NMR) is often a technique of choice for these studies, as it provides atomic-resolution information on structure, dynamics and interactions of IDPs. Nonetheless, NMR spectra of IDPs are typically extraordinary crowded, comparing to those of structured proteins. To overcome this problem, high-dimensional NMR experiments can be used, which allow for a better peak separation. In the present review different aspects of such experiments are discussed, from data acquisition and processing to analysis, focusing on experiments for resonance assignment.
New Article in Journal of Biomolecular NMR
Łukasz Jaremko, Mariusz Jaremko, Andrzej Ejchart, Michał Nowakowski

Simple and convenient method of protein dynamics evaluation from the insufficient experimental 15N relaxation data is presented basing on the ratios, products, and differences of longitudinal and transverse 15N relaxation rates obtained at a single magnetic field. Firstly, the proposed approach allows evaluating overall tumbling correlation time (nanosecond time scale). Next, local parameters of the model-free approach characterizing local mobility of backbone amide N–H vectors on two different time scales, S2 and Rex, can be elucidated. The generalized order parameter, S2, describes motions on the time scale faster than the overall tumbling correlation time (pico- to nanoseconds), while the chemical exchange term, Rex, identifies processes slower than the overall tumbling correlation time (micro- to milliseconds). Advantages and disadvantages of different methods of data handling are thoroughly discussed.
|
Sympozjum sekcji NMR Polskiego Towarzystwa Chemicznego 2018
New Article in International Journal of Biological Macromolecules
Michał Nowakowski, Łukasz Jaremko, Benedykt Wladyka, Grzegorz Dubin, Andrzej Ejchart, Paweł Mak

BacSp222 is a multifunctional bacteriocin produced by Staphylococcus pseudintermedius strain 222, an opportunistic pathogen of domestic animals. At micromolar concentrations, BacSp222 kills Gram-positive bacteria and is cytotoxic toward mammalian cells, while at nanomolar doses, it acts as an immunomodulatory factor, enhancing nitric oxide release in macrophage-like cell lines. The bacteriocin is a cationic, N-terminally formylated, 50-amino-acid-long linear peptide that is rich in tryptophan residues. In this study, the solution structure of BacSp222 was determined and compared to the currently known structures of similar bacteriocins. BacSp222 was isolated from a liquid culture medium in a uniformly 13C- and 15N-labeled form, and NMR data were collected. The structure was calculated based on NMR-derived constraints and consists of a rigid and tightly packed globular bundle of four alpha-helices separated by three short turns. Although the amino acid sequence of BacSp222 has no significant similarity to any known peptide or protein, a 3D structure similarity search indicates a close relation to other four-helix bundle-motif bacteriocins, such as aureocin A53, lacticin Q and enterocins 7A/7B. Assuming similar functions, biology, structure and physicochemical properties, we propose to distinguish the four-helix bundle bacteriocins as a new Type A in subclass IId of bacteriocins, containing linear, non-pediocin-like peptides.
|
|
|
|
|
|
Page 4 of 11 |



















